Figure 1.
Overview of the services that a cell bank should provide. A cell bank should offer various cell types from different species, such as primary cells, immortalized cell lines, and stem cells, tailored for specific applications. In addition, a cell bank should offer consultation, relevant information, and a range of support services to researchers.
STR profiling of HeLa cells. Two different panels of STR markers were used for the short tandem repeat profiling of HeLa cells from the Japanese Collection of Research Bioresources (JCRB). In panel (A), the STR panel is based on nine variant markers (D5S818, D13S317, D7S820, D16A539, vWA, TH01, Amelogenin, TPOX, and CSF1PO) which were amplified, and the sizes of the resulting amplicons were compared to ladders containing all known variant sizes. Panel (B) includes the eight markers D5S818, D13S317, D16A539, vWA, TH01, Amelogenin, TPOX, and CSF1PO from panel (A) along with seven additional markers (D3S1358, D21S11, D18S51, Penta E, Penta D, D8S1179, and FGA). This comparative analysis demonstrates the distinct allelic patterns generated by each profiling system, confirming the genetic identity of HeLa cells. The allelic patterns for HeLa cells are as follows: D5S818 (11,12), D13S317 (12,13.3), D7S820 (8,12), D16S539 (9,10), vWA (16,18), TH01 (7), Amelogenin (X), TPOX (8,12), CSF1PO (9,10), D3S1358 (15,18), D21S11 (27,28), D18S51 (16), Penta E (7,17), Penta D (8,15), D8S1179 (12,13), and FGA (18,21), respectively.
Figure 2.
STR profiling of HeLa cells. Two different panels of STR markers were used for the short tandem repeat profiling of HeLa cells from the Japanese Collection of Research Bioresources (JCRB). In panel (A), the STR panel is based on nine variant markers (D5S818, D13S317, D7S820, D16A539, vWA, TH01, Amelogenin, TPOX, and CSF1PO) which were amplified, and the sizes of the resulting amplicons were compared to ladders containing all known variant sizes. Panel (B) includes the eight markers D5S818, D13S317, D16A539, vWA, TH01, Amelogenin, TPOX, and CSF1PO from panel (A) along with seven additional markers (D3S1358, D21S11, D18S51, Penta E, Penta D, D8S1179, and FGA). This comparative analysis demonstrates the distinct allelic patterns generated by each profiling system, confirming the genetic identity of HeLa cells. The allelic patterns for HeLa cells are as follows: D5S818 (11,12), D13S317 (12,13.3), D7S820 (8,12), D16S539 (9,10), vWA (16,18), TH01 (7), Amelogenin (X), TPOX (8,12), CSF1PO (9,10), D3S1358 (15,18), D21S11 (27,28), D18S51 (16), Penta E (7,17), Penta D (8,15), D8S1179 (12,13), and FGA (18,21), respectively.
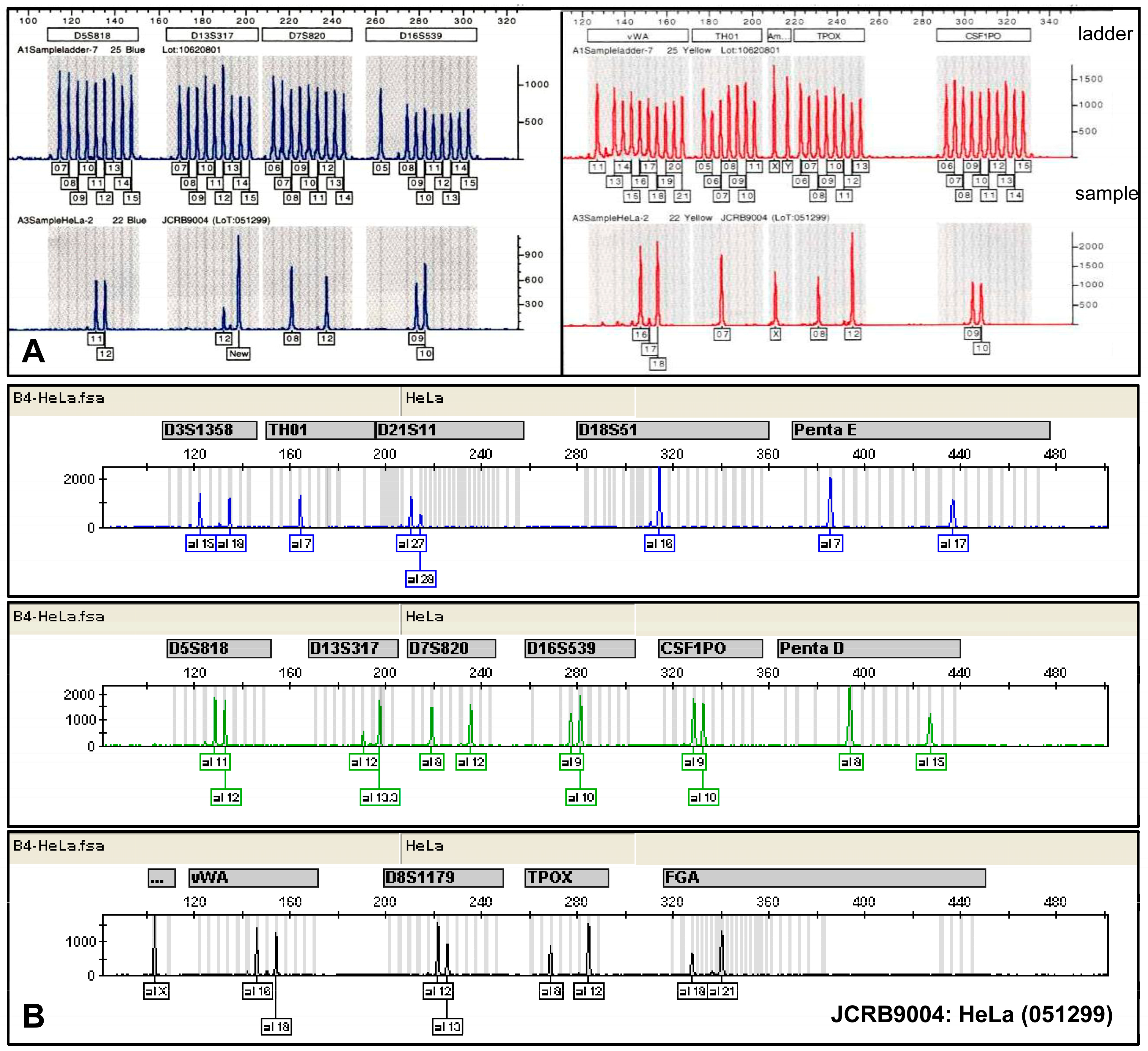
Figure 3.
Genetic characterization of the human transformed cell line known as Squeaky. Squeaky is a cell line similar to embryonic stem cells, obtained from the lung of a 14-week-old male fetus (MRC-5). This cell line was infected with a recombinant retrovirus that expressed the four factors Oct3/4, Sox2, Klf4, and c-Myc, resulting in an infinite lifespan. (A) G-banding analysis confirmed a male karyotype with a normal number of chromosomes, but an additional piece of genetic material was identified on chromosome 22 at the p11 region (marked with red arrow). (B) Further analysis using Multiplex Fluorescence In Situ Hybridization (m-FISH) revealed a derivative chromosome 22, which was a result of a translocation with chromosome 17, specifically affecting regions q21 and p11.2. This translocation can be best visualized using a whole chromosome probe for chromosome 17 (wcp17+).
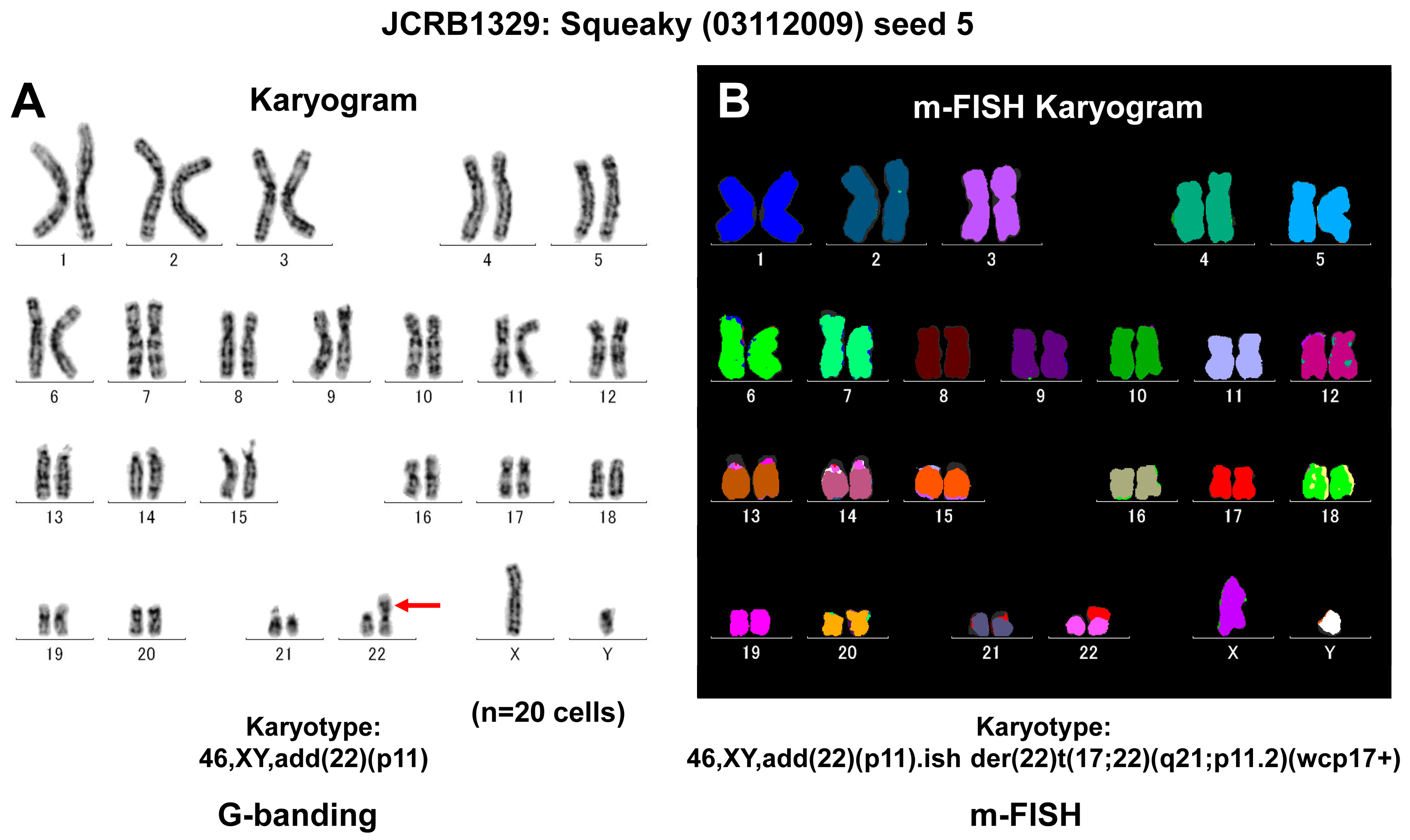
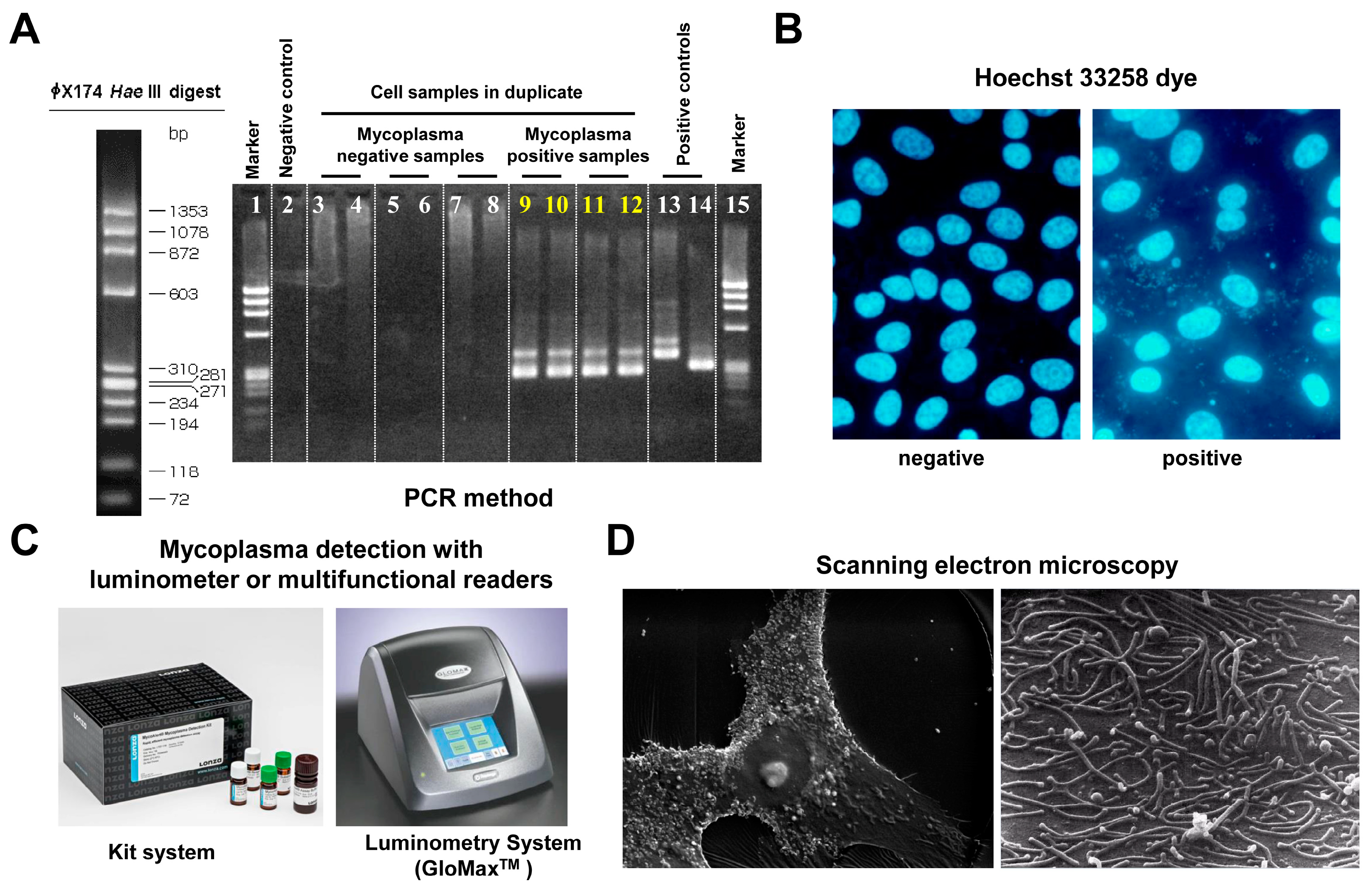
Figure 4.
Mycoplasma testing. There are several methods available for detecting mycoplasma contamination. (A) Mycoplasma detection by PCR involves amplifying specific DNA sequences unique to mycoplasma species, allowing for sensitive and rapid identification of contamination. In the depicted experiment, universal primers specific for most mycoplasma species were used. (B) Mycoplasma detection using the Hoechst 33258 bisbenzimide dye stain involves fluorescent staining of DNA, allowing for the visualization of mycoplasma cells under a fluorescence microscope. This method is advantageous because it can differentiate between eukaryotic and prokaryotic cells based on their size and morphology, providing a quick assessment of contamination in cell cultures. (C) The MycoAlert™ Mycoplasma Detection Kit is based on a selective biochemical test that exploits the activity of mycoplasma enzymes found in all six major mycoplasma cell culture contaminants and the vast majority of the 180 mycoplasma species. (D) Mycoplasma detection by scanning electron microscopy (SEM) allows for high-resolution imaging of mycoplasma cells, enabling observation of their unique morphology and attachment to host cells.
Multiplex PCR analysis of viral detection. (A) The setup of the multiplex real-time PCR system uses specific primers and probes to simultaneously amplify target sequences specific to DNA viruses (CMV, EBV, HHV-6, HHV-7, BKV, JCV, ADV, HBV, ParvoB19, and HPV18) as well as RNA viruses (HAV, HCV, HTLV-1, HTLV-2, HIV-1, and HIV-2). (B) The Epstein–Barr virus (EBV)-transformed lymphocyte cell line B95-8, which originates from Saguinus oedipus (cotton-top tamarin), was tested for the presence of EBV. The analysis confirms distinct amplification patterns indicative of EBV presence, with the housekeeping control GAPDH used as a reference; no other viruses were detected. (C) A standard sample (SD1) containing nucleic acids from various viruses underwent multiplex real-time PCR. This assay shows that the experimental setup can detect multiple viruses simultaneously. Further details of this assay can be found elsewhere [33]. The horizontal red arrow indicates the zero line, while the green horizontal arrow indicates the cut-off line. Panels (A,B) are screenshots from the multiplex real-time PCR system.
Multiplex PCR analysis of viral detection. (A) The setup of the multiplex real-time PCR system uses specific primers and probes to simultaneously amplify target sequences specific to DNA viruses (CMV, EBV, HHV-6, HHV-7, BKV, JCV, ADV, HBV, ParvoB19, and HPV18) as well as RNA viruses (HAV, HCV, HTLV-1, HTLV-2, HIV-1, and HIV-2). (B) The Epstein–Barr virus (EBV)-transformed lymphocyte cell line B95-8, which originates from Saguinus oedipus (cotton-top tamarin), was tested for the presence of EBV. The analysis confirms distinct amplification patterns indicative of EBV presence, with the housekeeping control GAPDH used as a reference; no other viruses were detected. (C) A standard sample (SD1) containing nucleic acids from various viruses underwent multiplex real-time PCR. This assay shows that the experimental setup can detect multiple viruses simultaneously. Further details of this assay can be found elsewhere [33]. The horizontal red arrow indicates the zero line, while the green horizontal arrow indicates the cut-off line. Panels (A,B) are screenshots from the multiplex real-time PCR system.
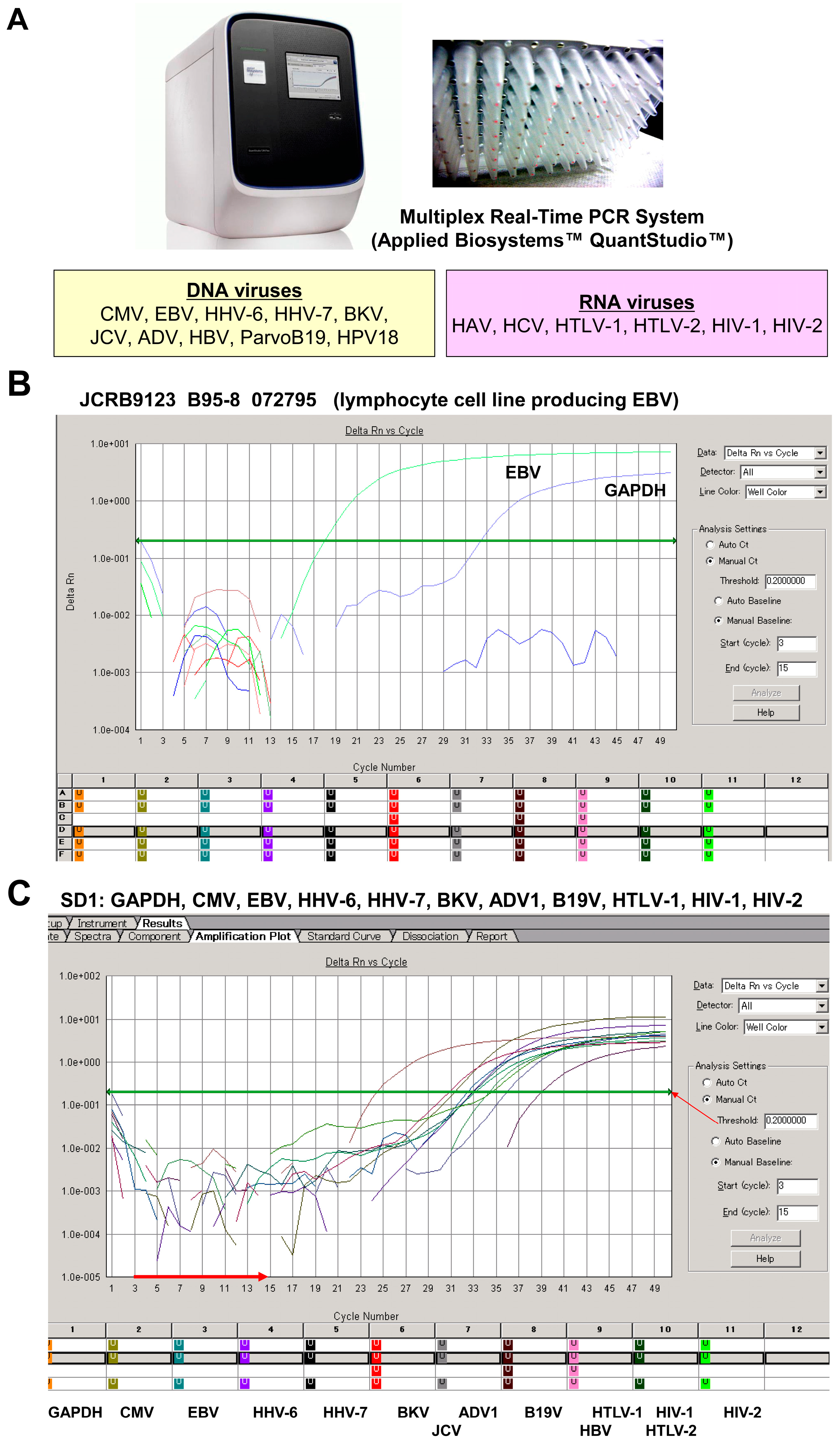
Figure 6.
Cell storage. The image depicts multiple liquid nitrogen (N2) tanks, each containing cryovials with frozen cells preserved at ultra-low temperatures. The frozen cells are stored in boxes within the vapor phase of the nitrogen, ensuring optimal conditions for maintaining viability and integrity over extended periods.

Figure 7.
Parceling of frozen cells for transport. (A) Various glass or plastic storage vials containing frozen cells prepared for dispatch are depicted. (B) A secure container designed to hold the vials during transit is shown. (C) The packaging process is highlighted, emphasizing the use of dry ice to maintain the low temperatures essential for preserving cell viability.

Figure 8.
Locations of representative cell culture biobanks worldwide. Depicted are various sites across America, Europe, Asia, and Australia where cell culture biobanks have been established. Each marked location represents a significant hub for cellular research and biobanking activities, contributing to advancements in biomedical science and regenerative medicine. The map highlights the global distribution of these facilities, emphasizing their importance in fostering international collaboration and innovation in the field of cell biology.
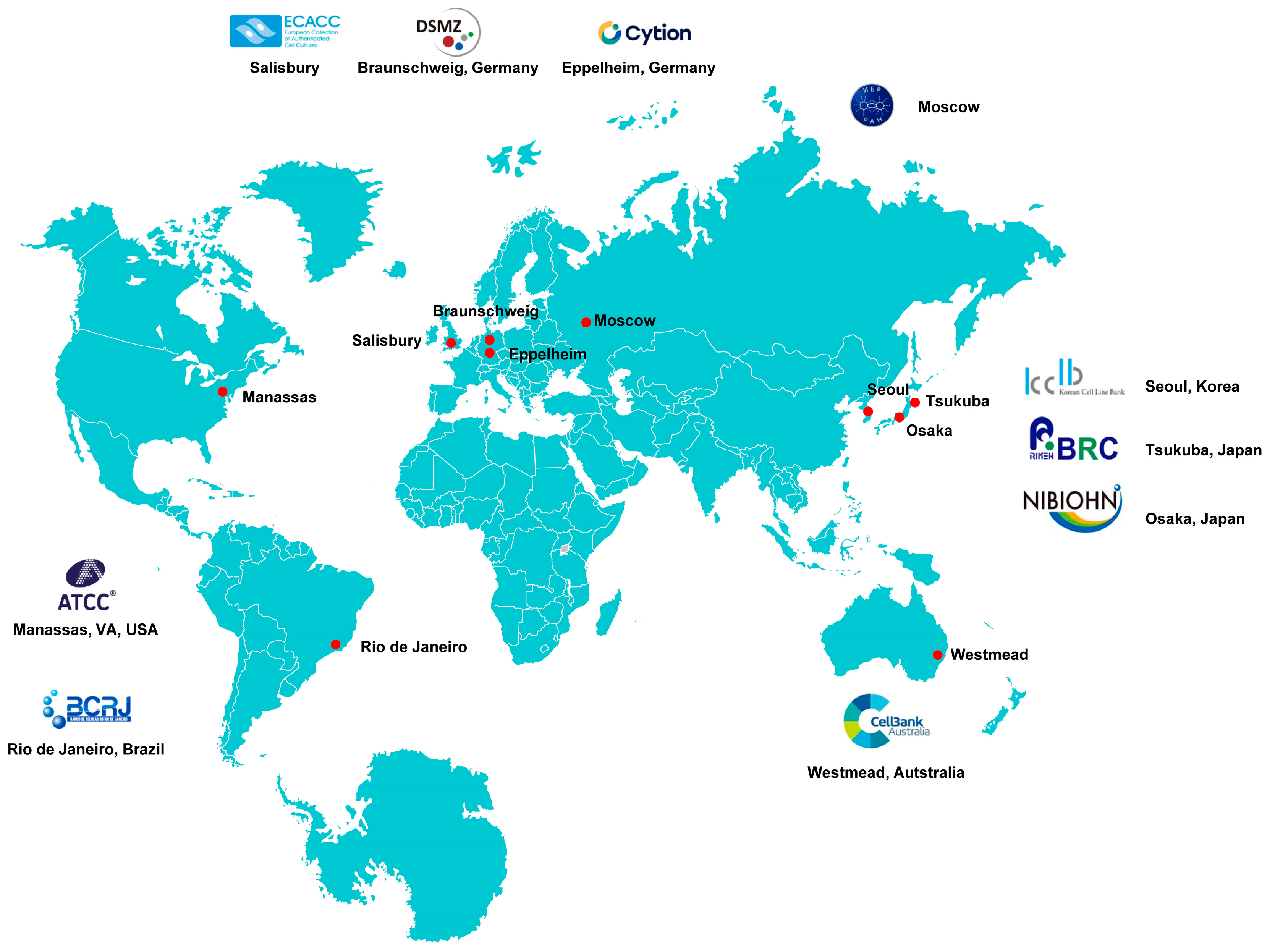
Table 1.
Summary of key information on selected cell bank repositories.
| Cell Bank Name | Location | Type of Cell Lines Offered | MTA | Quality Assurance Standards | Collaboration Opportunities | Contact Information 1 |
|---|---|---|---|---|---|---|
| ATCC | USA | >4000 human and animal cell lines | required | ISO 9001 [41] 2 ISO 13485 [42] 3, ISO/IEC 17025 [43] 4 ISO 17034 [44] 5 | Partnership with several companies and institutes (InSphero, USP, Synthgo, Qiagen, One Codex, NIST, and LGC Standards) | 10801 University Boulevard, Manassas, Virginia 20110-2209, USA Phone: (703) 365-2700 http://www.atcc.org 8 Email via contact form |
| ECACC | UK | >1100 cell lines from over 45 species | required | ISO 9001 [41] ISO/IEC 17025 [43] | Partnerships with many local distributors (Merck, KAC, and Cell Bank Australia) | UK Health Security Agency, Porton Down, Salisbury, SP4 0JG, UK Phone: +44 (0)1980 612512 https://www.culturecollections.org.uk/ 8 Email: [email protected] |
| JCRB 6 | Japan | 1642 cell lines, of which 1111 are of human origin | request and agreement form required | Testing for bacteria, fungi, mycoplasma, and viruses; performing species identification, cell identification, and chromosome analysis | Teamed up with the National Institute of Biomedical Innovation, Health and Nutrition (NIBIOHN) | 7-6-8 Saito-Asagi, Ibaraki Osaka 567-0085, Japan https://cellbank.nibiohn.go.jp/english 8 Email: [email protected] |
| DSMZ | Germany | >900 from various species | required | Testing for growth characteristics, bacteria (notably mycoplasms), fungi, yeast, and human pathogenic viruses; species identification and authentication | Member of several national and international organizations, networks, and projects | Inhoffenstraße 7B 38124 Braunschweig Science Campus Braunschweig-Süd, Germany Phone: +49 (0)531 2616-0 https://www.dsmz.de/ 8 Email via contact form |
| KCLB | South Korea | 423 cell lines from various human tissues (gastric, colon, lung, cervical, ovarian, pancreas, breast, and other cancer cell lines) | required | Scientific quality control including STR profiles | Contract research organization service, hands-on workshops | Cancer Research Institute, Seoul National University College of Medicine, 103, Daehak-ro, Jongno-gu, Seoul, Republic of Korea, 03080 Phone: +82-02-3668-7915 Email: [email protected] https://cellbank.snu.ac.kr/eng/ 8 |
| BCRJ 7 | Brazil | Primary cells and immortalized cell lines | NN | Several services including cell storage, screening for mycoplasma, toxicity tests, cell immortalization, and cell authentication | Offers courses in basic and good practices in cell culture | Av. N. S. das Gracas, 50, Prédio 32, Parque Tecnológica de Xerém Duque de Caxias, Rio de Janeiro, Brazil Phone: +55 21 2145-3337 https://bcrj.org.br/ 8 [email protected] |
| Cytion | Germany | >800 human and animal cells, stem cells, and primary cells | not required | ISO 9001 [41] Mycoplasma testing via colorimetric assay and a PCR-based method, STR analysis, and testing for viral/bacteria/fungi contaminants | Collaborations with industry and academic institutions | CLS Cell Lines Service GmbH Dr.-Eckener-Str. 8 69214 Eppelheim, Germany https://www.cytion.com/ 8 Phone: +49 (0)6221 405780 Email: [email protected] |
| RCCC | Russia | ~150 mammalian cell lines (mouse, dog, rabbit, rat, monkey, pig, and hamster; human primary and cancer cells | NN | Cells are tested for contamination; STR profile | Associated with the Koltzov Institute of Development Biology which offers Doctoral/PhD programs | Koltzov Institute of Developmental Biology of the Russian Academy of Sciences 26 Vavilov Street, Moscow Phone: +7 (499) 135-33-22 Email via contact form |
| RIKEN BRC | Japan | NN | required, some cells with restrictions | ISO 9001 [41] Provides validation reports on request; started with cell verification testing service | Provides annual technical training course for researchers, students, and technicians; offers cooperation (e.g., deposition of cells) | Riken BioResource Center 3-1-1 Koyadai, Tsukuba, Ibaraki, 305-0074, Japan https://cell.brc.riken.jp/en/ 8 Email: [email protected] |
| CellBank Australia | Australia | >1300; distributes cell lines from ECACC and JCRB. In addition, several mouse and human cancer cell lines are distributed | required | ISO 9001 [41] ISO/IEC 17025 [43] | Partnership with ECACC and JCRB | Children’s Medical Research Institute 214 Hawkesbury Road Westmead NSW 2145 Locked Bag 23 Phone: +612 8865 2850 https://www.cellbankaustralia.com/ 8 Email: [email protected] |
Source link
Sabine Weiskirchen www.mdpi.com