1. Introduction
Monitoring the fetal heart rate during pregnancy is a crucial diagnostic tool for assessing the in utero condition of the fetus, enabling the timely detection of fetal distress, arrhythmias, acidosis, and certain pathological conditions related to fetal hypoxia [
1]. Fetal heart rate evaluation during the perinatal period is one of the primary measures to reduce pregnancy-related risks and enhance the quality of fetal outcomes [
2].
Currently, mainstream fetal heart rate monitors are categorized into three types based on their operational principles: fetal phonocardiography, fetal cardiotocography, and fetal electrocardiography [
3]. The fetal QRS complex, observed in fetal electrocardiograms, represents the depolarization of the fetal ventricles, which consists of Q, R, and S waves. Fetal electrocardiography involves placing electrodes on the fetus’s head or the maternal abdomen to obtain the QRS complex that characterizes fetal cardiac activity, divided into invasive and non-invasive detection methods. The non-invasive approach, which directly uses the maternal abdominal electrical signals as the measurement signal, is entirely non-invasive, more readily accepted by pregnant women subjectively, and can be used for long-term monitoring. However, separating the fetal ECG signal can be challenging due to interference from other physiological signals and noise [
4], such as maternal ECG, baseline drift, and electromyographic interference [
5]. The amplitude of the maternal ECG signal is significantly larger than that of the fetal ECG signal, and there is also spectral overlap [
6]. Therefore, to obtain a reliable fetal ECG signal, it is crucial and necessary to eliminate the interference of the maternal ECG.
To address the challenges of fetal heart rate extraction, several mathematical methods have been proposed, including blind source separation algorithms based on Independent Component Analysis (ICA) and Singular Value Decomposition (SVD) [
7], algorithms combining power spectral density and matched filtering techniques [
8], adaptive filtering algorithms combining SVD and smooth window methods [
9], wavelet separation methods combining clustering algorithms with extrema pairs and ICA [
10], and a negative entropy-based blind source separation method combined with a template subtraction method [
11]. Kahankova et al. proposed a method combining ICA and LMS, achieving Se, PPV, and F1 scores of 89.41%, 90.42%, and 89.19%, respectively [
12]. Mansourian et al. proposed a method for extracting fetal QRS complexes from single-channel abdominal ECG signals based on an Adaptive Improved Permutation Entropy (AIPE) algorithm, achieving an F1 score and Se of 90.95% and 90.77%, respectively [
13]. Other researchers have used methods such as Principal Component Analysis [
14,
15], Recursive Least Squares [
16], Tensor Decomposition [
17], and Linear Combination Algorithms [
18] for fetal heart rate detection research, all achieving satisfactory experimental outcomes.
With significant advancements in deep learning technologies, numerous studies have used neural networks to extract fetal ECG signals without separating the maternal ECG [
19,
20,
21]. Nguyen et al. proposed a novel method based on Conditional Generative Adversarial Networks (cGANs) for extracting fetal electrocardiogram (ECG) signals in the time–frequency domain using the Short-Time Fourier Transform (STFT) method. This method demonstrates higher robustness and resistance to interference compared to traditional approaches [
22]. Zhong et al. employed a one-dimensional convolutional neural network (CNN) to detect fetal QRS complexes without removing the maternal ECG signal, aiming for a similar goal as this study, achieving an accuracy of 75.33%, a recall rate of 80.54%, and an F1 score of 77.85% [
23]. Lee et al. proposed a more profound CNN architecture that utilizes four-channel original ECG signals to detect fetal QRS, resulting in a positive predictive value (PPV) of 92.77% and an average sensitivity of 89.06% [
24]. Vo et al. used the Short-Time Fourier Transform (STFT) method to prepare features for training, employing ResNet and a one-dimensional octave convolution network to design a four-class deep learning model, which achieved an accuracy of 91.1% [
25].
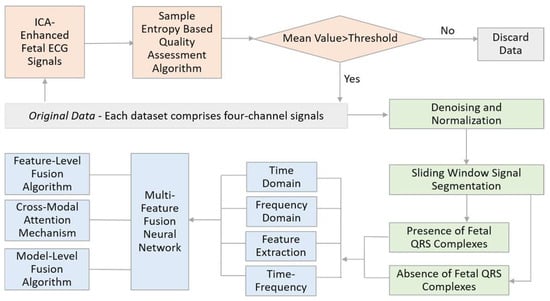
This study introduces a non-invasive fetal QRS complex recognition method based on a multi-feature fusion neural network. Initially, we designed a signal entropy data quality detection algorithm based on blind source separation to ensure the selected maternal ECG signals meet quality requirements. Subsequently, the signals were subjected to data preprocessing operations such as denoising and normalization to prepare for further analysis. Following this, the maternal ECG signals were segmented using a sliding window technique and computed across four modalities: time domain, frequency domain, time–frequency domain, and nonlinear characteristic values. Ultimately, we proposed three types of deep neural networks that integrate multiple features to identify fetal QRS complexes rapidly. The overall algorithmic workflow of this topic is illustrated in
Figure 1.
The method proposed in this paper has four main advantages. First, traditional methods require at least four channels to separate fetal ECG signals from maternal ECG signals, which can be uncomfortable for pregnant women in clinical applications and have significant limitations. The method used in this paper does not require the separation of maternal and fetal ECG signals. It can directly detect fetal heart QRS complexes from a single-channel fetal ECG signal obtained from the abdomen of pregnant women. Second, fetal ECG waveforms can intuitively display the fetal cardiac activity status, containing valuable information for doctors to diagnose and treat. Traditional methods can only calculate the fetal heart rate and fetal heart rate variability, whereas the technique proposed in this paper can extract complete fetal ECG waveforms with apparent features. This allows for the precise prenatal monitoring of fetal health based on morphological information from the fetal ECG. Third, this paper explores lightweight networks with lower storage and computation requirements. By using a window moving method, it is possible to quickly label the position and shape of the fetal heart, achieving the real-time monitoring of the fetal ECG. Fourth, including multimodal data provides the deep learning model with more prosperous and adequate information. The model’s overall performance is improved by leveraging the complementarity of information across multiple modes.
3. Multi-Feature Fusion Neural Networks
Numerous scholars have utilized CNNs to extract fetal ECG signals from pregnant women [
33,
34,
35,
45]. However, these algorithms exhibit certain limitations. Firstly, although CNNs can autonomously learn effective features from raw ECG data, these features often need more interpretability. Moreover, some essential features that can be easily extracted manually are challenging to learn through CNNs. Secondly, scholars tend to employ multiple layers of complex neural networks to enhance the network’s recognition accuracy, resulting in slower computational speeds and higher demands on hardware resources, thereby limiting the practical application of these advancements.
Therefore, developing multi-feature fusion neural networks that are both highly accurate and cost-efficient in computation holds significant importance. Such networks are better equipped to adapt to data distributions and task requirements. Common multimodal fusion strategies are primarily categorized into early fusion, late fusion, and multi-stage fusion. Early fusion refers to integrating data from different modalities into a single feature representation during the initial feature extraction phase. Late fusion involves the integration of results after features from different modalities have been extracted and processed independently. Multi-stage fusion strategies integrate information from multiple modalities at various stages. This paper designs three distinct model fusion networks and compares the accuracy and merits of three fetal ECG classification algorithms that fuse features across multiple domains.
3.1. Feature-Level Fusion
Feature-level or early fusion is the most commonly used strategy in multimodal recognition systems. It involves the direct combination of time-domain, frequency-domain, time–frequency domain, and nonlinear features obtained after preprocessing into a 27 × 60 matrix. This matrix is then fed into a neural network for learning and classification. The network diagram for feature-level fusion is shown in
Figure 4. This study employs Resnet34 to learn the early fusion of multimodal data, with a training duration of 50 rounds [
46]. ResNet34 efficiently captures complex patterns from multimodal data, achieving a high classification accuracy through the early fusion of diverse feature types while maintaining computational efficiency.
3.2. Late Fusion
The cross-modal attention mechanism dynamically allocates different weights to each modality by learning the correlations between modalities, thus better-integrating information for specific tasks. This study utilizes a multimodal attention model that accepts four types of input data: time-domain data processed by a Long Short-Term Memory (LSTM) model, frequency-domain data handled by a 1D-CNN model, nonlinear feature data analyzed by an FCN model, and all feature data processed by a ShuffleNet model. Initially, each model processes its input data, which are then concatenated along the feature dimension. Through a multi-head attention layer, features are integrated and weighted for attention, facilitating the interaction and integration of information between different features [
47,
48]. The final classification result is output through a fully connected (FC) layer with an output dimension of two.
Using LSTM for Time-Domain Signals: ECG signals are time-domain signals, making LSTM networks suitable for processing them [
49]. The LSTM model used in this study consists of an LSTM layer and an FC layer, designed to process sequential signals with an input dimension of 1 × 60. The output of this network is four feature values.
Using 1D-CNN for Frequency-Domain Signals: The 1D-CNN network used here includes two convolutional layers, ReLU activation functions, a max pooling layer, and two FC layers, effectively extracting and classifying local features from frequency-domain data [
50]. The input to this network is sixty values, and the output is four values.
Using DNN for Feature Value Signals: Given the small amount of feature value data extracted from each segment of the ECG signal, this study uses an FC neural network model to analyze feature value signals. The network consists of four FC layers, each followed by a ReLU activation function, automatically extracting valuable features for classification tasks. The input to this network is sixty values, and the output is four values.
Using ShuffleNet for All Modal Data: ShuffleNet is a lightweight deep neural network architecture designed for efficient computation and parameter usage [
51]. Given that the time–frequency domain signals are 24 × 60, significantly larger than the signals from the first three modalities, ShuffleNet was chosen for its ability to quickly analyze such data. The output of this network is four feature values.
The network diagram for late fusion based on the cross-modal attention algorithm is shown in
Figure 5. Since the LSTM network is suitable for processing time-domain signals, it can capture the temporal dependencies within ECG signals. The 1D-CNN can efficiently extract local features from frequency-domain signals. The DNN model can automatically extract valuable information from a small amount of feature value data. ShuffleNet, due to its lightweight design, can quickly process larger size time–frequency domain signals. Finally, the multimodal attention model can comprehensively utilize the characteristics of various types of data, achieving the efficient and accurate classification of ECG signals. The combination of these specialized networks, alongside the cross-modal attention mechanism, allows the model to dynamically allocate computational resources based on the importance of different features. This design ensures that high classification accuracy is achieved without excessive computational overhead, striking a balance between accuracy and efficiency.
3.3. Model-Level Fusion Algorithm
The Multi-Layer LSTM (ML-LSTM) method is a model-level fusion method that integrates different modal data at various stages of the network, enabling the acquisition of a joint feature representation of four types of characteristics from the pregnant woman’s ECG signal for the recognition of fetal ECG QRS complexes [
49].
First, the time-domain features are fed into the first layer of the LSTM network, with an input dimension of 60 and a hidden unit size of 64. Then, the frequency-domain features are concatenated with the hidden state outputs of the first-layer LSTM (64 dimensions) and input into the second-layer LSTM, with an input dimension of 65 (64 + 1) and a hidden unit size of 64. Next, nonlinear features are concatenated with the hidden state outputs of the second-layer LSTM (64 dimensions) and introduced into the third-layer LSTM, with an input dimension of 65 (64 + 1) and a hidden unit size of 64. Finally, the hidden state of the third-layer LSTM (64 dimensions) is concatenated with the time–frequency domain features (24 dimensions) and fed into the fourth-layer LSTM, with an input dimension of 89 (64 + 25) and a hidden unit size of 64. In this way, the fourth-layer LSTM can simultaneously process information from the time domain, frequency domain, time–frequency domain, and nonlinear features. These features are then fed into a fully connected layer (FC layer) for the final classification task, generating the ultimate prediction results. The network architecture of the model-level fusion algorithm based on ML-LSTM is illustrated in
Figure 6.
By progressively combining different types of features at each LSTM layer, the ML-LSTM model efficiently captures the complex relationships between various signal characteristics. This layered fusion approach not only enhances classification accuracy but also optimizes resource allocation, ensuring that the computational load remains manageable.
4. Results
4.1. Model Training and Classification
The Cosine Annealing Learning Rate Scheduler is a commonly employed strategy for adjusting the learning rate in the training process of neural networks. In this study, the Cosine Annealing Learning Rate Scheduler was applied, and the learning rate was decreased from an initial value of 0.001 to a final value of 0.0001. The dataset was divided into non-overlapping training and test sets, with a ratio of 7:3, resulting in a training set of 65,408 and a test set of 28,033 entries.
This study utilized the entire ADFECGDB dataset as the test set to validate the accuracy and reliability of the proposed algorithm. Each data entry consisted of 1 min recordings, with a total of 11,492 entries in the dataset.
4.2. Evaluation Metrics
This study employs accuracy, sensitivity, specificity, PPV, F1 score, and the ROC curve to measure the performance of the classification model. Accuracy refers to the ratio of the number of correctly predicted samples to the total number of samples; sensitivity, also known as recall, indicates the model’s ability to identify positive cases correctly; specificity refers to the model’s ability to identify negative cases correctly; PPV represents the proportion of samples predicted as positive that are positive; the F1 score considers both the precision and recall of the model, representing their harmonic mean. The ROC curve is a graphical tool used to evaluate the performance of binary classification models. The area under the ROC curve (AUC) is often used to quantify the model’s overall performance. An AUC value closer to 1 indicates better model performance and a more remarkable ability to distinguish between positive and negative samples.
4.3. Results and Discussion
Table 1 presents a comparative analysis of experimental results from various research methods, comparing the multimodal and unimodal methods proposed in this paper, ablation studies, and other research.
Figure 7a illustrates the accuracy trend of different models during training. It can be clearly observed that the feature-level fusion method based on ResNet34 converges faster and achieves a higher final accuracy compared to other methods. Meanwhile,
Figure 7b presents the ROC curve for the ResNet34 feature-level fusion model, with an area under the curve (AUC) of 0.99, indicating excellent classification performance. These results further validate the robustness of the proposed method in handling multimodal data.
From
Table 1 and the figures, it is evident that multimodal methods, especially the ResNet34-feature level fusion, significantly outperform unimodal methods in all evaluation metrics. The ResNet34-feature level fusion achieved a 95.85% accuracy, 97% sensitivity, 95% specificity, and an F1 score of 91%, demonstrating its strong overall performance. In comparison, the unimodal methods, such as LSTM in the time domain and CNN in the frequency domain, show noticeably lower results, with accuracy values of 85.42% and 88.41%, respectively. The fusion approach effectively leverages the complementary information among various features, thereby enhancing classification performance.
Moreover, the results of testing the ResNet34-feature level fusion algorithm on the ADFECGDB dataset are also impressive, achieving a high accuracy of 97.89%, with a sensitivity of 97% and a specificity of 98%. This indicates that the proposed method not only demonstrates high reliability but also possesses transferability, as it performs effectively across different datasets, validating its potential for broader applications in fetal heart monitoring.
The ablation study further validates the importance of each feature to the model’s performance. Removing the time–frequency domain features has the most significant impact on performance, indicating their crucial role in fetal heart signal classification. In contrast, removing time-domain, frequency-domain, and feature values has a relatively minor impact. A comparison with other studies shows that the methods proposed in this paper generally outperform others in various performance metrics, with all compared studies utilizing the same database, the PhysioNet/CinC Challenge 2013 dataset.
Overall, the multimodal methods proposed in this paper demonstrate strong performance and wide applicability in the classification of fetal heart ECG signals. The comprehensive analysis across multiple databases and the ablation study further validate the robustness of the proposed methods, providing important references for their potential application in clinical and wearable health monitoring systems.
5. Conclusions
High-quality fetal ECG signals are of significant clinical importance for diagnosing the health status of fetuses. Our method leverages a sliding window approach to identify fetal QRS complexes from maternal ECG signals sequentially while employing a lightweight deep neural network for the rapid and accurate output of fetal QRS complex waveforms, paving the way for the intelligent morphological analysis of fetal ECG signals. This approach overcomes the limitations of traditional fetal heart rate monitoring, which requires the separation of maternal and fetal ECG signals. Compared to conventional methods that necessitate four-channel detection, our method achieves the reliable detection performance of fetal QRS complexes with data from only a single channel under the premise of ensuring data quality. This means that pregnant women, using a single electrode patch attached to the abdomen, can independently monitor the fetal heartbeat status at home using a smartphone, presenting a broad application prospect.
However, the current dataset does not include cases of high-risk pregnancies or multiple pregnancies, which are important scenarios for clinical practice. Future research should focus on adapting fetal heart rate detection methods to effectively function in such challenging conditions. Looking forward, the further training of the network with more extensive and diverse datasets is necessary to enhance analysis accuracy and robustness. Future improvements could also involve integrating advanced noise reduction techniques and adaptive algorithms that account for different maternal conditions and environments, thereby increasing the method’s applicability and reliability in clinical practice.