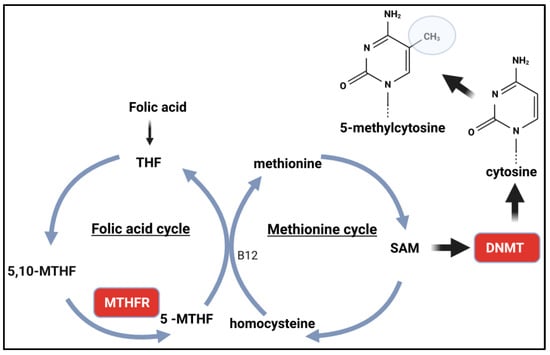
Polymorphisms in the MTHFR Gene and Global DNA Methylation
The polymorphisms in the MTHFR gene that were analysed with regard to their association with the global methylation profile are listed in Table 1. These polymorphisms are single nucleotide polymorphisms (SNP) found in the promoter, exons, introns and 3′-UTR (Untranslated Region) regions, and some of them have a previously unknown function.
Table 2 shows the extraction of data from the articles, which are listed in order of publication [19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51]. The most analysed polymorphism is rs1801133 (C677T). This single nucleotide polymorphism causes the replacement of Ala222Val, is related to the T allele and leads to the formation of a thermolabile molecule with a loss of ~70% of catalytic activity [52].
When analysing exposure to benzene for its carcinogenic potential, it has been shown that carriers of the rs1801133 TT genotype tend to have lower levels of global methylation and greater induction of micronuclei compared to carriers of the rs1801133 C allele [41]. This finding may be related to folate metabolism, which, when impaired, reduces the conversion of uracil to thymine, leading to misincorporation of uracil into DNA and consequently favouring chromosomal breaks [53], which could lead to an important mechanism favouring cancer [17].
As expected, an association between the homozygous rs1801133 TT genotype and lower levels of global methylation was found in studies with different designs, such as in samples from Russian blood donors [33]. Other studies have demonstrated the association of rs1801133 T with the methylation level of LINE-1 in samples from different clinical conditions. In patients with schizophrenia, no association between LINE-1 methylation and the rs1801133TT genotype was observed in males, while lower global methylation was found in females and the rs1801133TT genotype, even when folate levels were controlled [27]. LINE-1 methylation levels were also significantly lower in patients carrying the T allele with or without coronary atherosclerosis [37]. When analysing patients diagnosed with hypertension with and without medication, the TT genotype was only associated with lower methylation levels in patients without medication [48]. Another study looking at medication use showed that lower levels of global methylation were associated with the rs1801133 TT genotype in patients with recurrent glioblastoma who were or were not treated with intranasal perillyl alcohol [44]. Common to these studies is that the effect of the SNP appeared to be associated with variables such as gender [27] or medication [44,48]. The data from studies with medication suggest that the ability of a drug to alter the methylation profile needs to be better explored. Indeed, many drugs have been tested for this purpose, but little is known about the drugs that are already widely used [54].
Following this line of reasoning, two studies analysing peripheral blood found lower levels of global methylation in samples from mothers of Croatian Down syndrome carriers in the LINE-1 sequences [35] and from Brazilians in the ALU sequences [49]. While this result in the Croatian study was associated with a lower estimated folic acid consumption and the rs1801133 T allele [35], the Brazilian study showed a higher folic acid level and a lower level of global methylation without influence of the rs1801133 genotype on the result. Here, the comparison is between two studies with the same clinical condition but differing in experimental design. While Mendes et al. [49] collected data from postpartum women at the first postnatal consultation, Božović et al. [35] do not clarify the timing of recruitment. While the Croatian study estimates folic acid intake based on the consumption of certain foods, Mendes et al. use the blood dose of this metabolite. The different analytical approaches may help to explain the discrepant results on the modifying effect of folic acid levels and genotypes. However, it is also necessary to take into account the complexity of the metabolic pathway involved, the necessary and unquantified cofactors and the different genetic backgrounds. Regarding the complex pathway, a study of pregnant women in the first trimester showed an association of the T allele with lower peripheral blood methylation levels only in mothers with vitamin B6 deficiency [22].
It seems at least partially plausible to hypothesise that the rs1801133 genotype influences MTHFR activity but that clinical, environmental, nutritional, genetic and other unforeseen factors may also influence this relationship. One way to establish this relationship would be through intervention studies, which are currently scarce in the literature. An experimental approach in humans has shown that high-dose folic acid (5 mg/dL) has global effects on the sperm methylome of infertile men [34]. Reduced representation bisulfite sequencing (RRBS), which encompassed the methylation status of ∼1.8 million CpGs, revealed a significant loss of methylation in intergenic regions, introns and exons, and the observed response was quite different among MTHFR genotypes. Global methylation loss in sperm of MTHFR 677 CC males was significant only in intergenic regions, while the presence of at least one T allele (CT or TT) was associated with significant methylation loss in all sequenced tiles in promoters, exons, introns and intergenic regions. According to the authors, possible regulatory effects could occur that impair the MTHFR activity of carriers of the TT genotype and reduce the availability of methyl groups from folic acid, which would impair the cellular methylation capacity. In a zebrafish mthfr model (mimicking the rs1801133CT or TT genotype), the effect of folic acid supplementation, in addition to reducing the SAM:SAH ratio, showed a relevant increase in cystathionine concentration that was not observed in the mthfr control, which could explain the effects [55].
The lack of correlation between the rs1801133 TT genotype and DNA methylation levels was also observed in other experimental designs. A subsample of the ‘Healthy Survey of São Paulo study, representative of the population of São Paulo/Brazil, analysing food consumption, plasma markers of one-carbon metabolism and global methylation and polymorphisms of interest, revealed an age-dependent variation in methylation levels, with younger individuals showing lower levels than adults and the elderly. However, no correlation was found when rs1801133 genotype groups were compared [47]. In samples from healthy vegetarian Indian individuals, no association was also found between rs1801133 genotypes and methylation levels regardless of age group and vitamin B12 status [50]. Other studies conducted with blood or oral mucosal samples from healthy populations, including studies of adolescents, adults, men and women with their newborns, also failed to detect associations of global and LINE-1 methylation with rs1801133 [21,23,24,28,29]. In clinical tumour diseases such as colorectal cancer, breast cancer and acute lymphoblastic leukaemia, there is also no correlation between genotype and methylation [20,26,43,51]. In the study of children with acute lymphoblastic leukaemia, the outcome of oral mucositis was associated with different levels of methylation, suggesting hypomethylation in the mucosal recovery phase, although the MTHFR gene polymorphism had no effect [51]. In studies looking at colorectal cancer and breast cancer, there was no association between tumour and global and LINE-1 methylation in peripheral blood [20,26,43]. It is possible that studies using samples taken directly from the tumour may show some correlation, as changes in the DNA methylation profile are one of the hallmarks of cancer [56]. In other diseases, such as autoimmune diseases, cerebrovascular diseases and dementia, no associations between global methylation and rs1801133 genotype or clinical condition and global methylation have been observed either [25,31,39]. In a study of patients with diabetes, no association was found between methylation and genotype. However, higher methylation levels were observed in untreated diabetic patients than in treated patients, suggesting that treatment may influence this profile [38].
Surprisingly, there are also reports of a correlation between variants with lower MTHFR activity and increased global methylation. In healthy women, systemic inflammation, indicated by higher serum C-reactive protein, is weakly associated with global DNA hypermethylation of leukocytes in peripheral blood. This association was more evident in individuals carrying the minor allele of MTHFR rs1801133 [40]. Another study in which samples from pregnant women with pre-eclampsia were analysed in comparison to healthy pregnant women showed that cases carrying the CT genotype rs1801133 had significantly higher global DNA methylation than CT-matched controls. According to the authors, this suggests that women carrying heterozygotes with hyperglobal DNA methylation are more susceptible to pre-eclampsia [42]. It is worth noting that in the study conducted with pregnant women, the rs1801133 genotype was not analysed, as there were no controls in this genotype group [42]. Along the same lines, but outside the inflammatory context, are results with samples from various studies conducted in the UK, which also indicate a link between the rs1801133 TT genotype and higher levels of global methylation. In this study, intervention with riboflavin was able to reduce the previously elevated homocysteine levels in this test group but had no effect on LINE-1 hypermethylation [46]. In relation to these results, it is important to emphasise that in the studies by Nojima et al. [40] and Mishra et al. [42], a chronic inflammatory state is present, as pre-eclampsia is strongly associated with an increase in inflammatory markers, including C-reactive protein [57]. In the study by Amenyah et al. [46], the description of the population is not clear enough in terms of inclusion criteria, but the rs1801133 TT group has a higher percentage of hypertensives and higher homocysteine levels, a biomarker associated with markers of inflammatory factors including C-reactive protein [58]. This profile was also found in tumour samples and adjacent non-neoplastic skin from kidney transplant patients [19]. Kidney transplantation involves a number of pathophysiological changes from donor to recipient, and studies have discussed the presence of subclinical inflammation in these patients [59]. Given this background, it can be speculated that the global hypermethylation effect appears to be associated with a pattern that favours inflammation. Furthermore, this finding is independent of the method used to analyse global methylation, as it occurred in studies where the method was ELISA [42], luminometric [40] or pyrosequencing of LINE regions [19,46].
The rs1801131 also affects MTHFR activity but to a lesser extent than the rs1801133. rs1801131 causes the Glu429Ala (A1298C) mutation associated with the C allele and has a less significant effect on enzymatic activity [60].
In breast tissue samples from healthy women, carriers of the rs1801131 C allele showed 4% lower LINE-1 methylation [36]. Samples from a group of patients with non-syndromic orofacial clefts showed lower levels of global methylation in oral mucosa samples than the control group. However, rs1801131 CC was associated with lower LINE-1 methylation levels in the control group but not in the cases [45]. On the other hand, the rs1801131 C allele was associated with a higher percentage of methylation in DNA samples extracted from leucocytes of prehypertensive and hypercholesterolaemic individuals than the group carrying the AA genotype [30]. Similarly, the AC genotype is associated with higher methylation levels in the LINE-1 regions in the peripheral blood of patients with colorectal cancer with advanced clinical and pathological stages and aged over 61 years [43]. It is noteworthy that the two studies presenting results in which the rs1801131 CC genotype is associated with lower global methylation levels did not analyse samples of leukocyte DNA but of breast [36] and oral cavity [45]. None of these studies observed the effects of the rs1801133 genotype on global methylation levels.
As with rs1801133, no correlation was found between the rs1801131 polymorphism and DNA methylation levels in other experimental designs. In a case-control study comparing blood samples from patients with and without a breast cancer diagnosis, the rs1801131 genotype had no effect on the level of LINE-1 methylation, which in turn was lower in the clinical samples of the case group [32]. Other studies using samples of peripheral blood or oral mucosa from patients with colorectal cancer, breast cancer, acute lymphoblastic leukaemia, autoimmune diseases, cerebrovascular diseases, benzene-exposed workers, mothers of children with Down syndrome and healthy individuals also found no association with global, LINE-1 or Alu methylation [20,22,24,25,26,31,40,41,47,49,51].
Two studies analysed further SNPs in addition to rs180133 and rs180131. Wernimont et al. [21] investigated the association of methylation in the LINE-1 and Alu regions with the SNPs rs12121543, rs13306556, rs1537516, rs17367629, rs17421462, rs1994798, rs3737965, rs4846049 and rs6541003 in the peripheral blood of healthy men and found no association. Deroo et al. [32] analysed the SNPs rs3737967 and rs1537514 in the peripheral blood of healthy women with sisters who had breast cancer, and rs1537514 was associated with increased LINE-1 methylation and a lower risk of breast cancer. However, the authors do not clarify which allele or genotype is associated with this profile. This SNP is located in the 3′-untranslated region, and predictions in SNPinfo indicate that it is a putative microRNA binding site, suggesting that these polymorphisms are associated with changes in the expression of MTHFR.
In summary, an intuitive conclusion based on the functionality of SNPs in MTHFR suggests that reduced enzyme activity (or reduced expression) could lead to reduced SAM levels and, consequently, reduced DNA methylation. Indeed, some studies show this, but others show a correlation with higher methylation levels. The data from these studies show that other factors may contribute to this profile or predominate over genotype. Other studies have also shown that the intuitive association depends on a reduction in the levels of certain substrates that act in the folic acid cycle.
Source link
Naila Francis Paulo de Oliveira www.mdpi.com