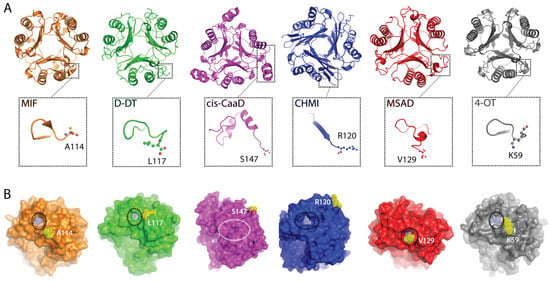
2.1. Crystallographic Analysis of TSF Representative Members Reveals Noticeable Differences in the C-Terminal Region
Previously published studies on the TSF human members, MIF and D-DT, showed that the C-terminal region has a multi-tiered role associated with the structure and function of these proteins [14,16,17,34,35,36]. Having these two proteins as our benchmark, we probed for broader insights into this superfamily. While interrogation of over 11,000 proteins is unrealistic, we selected representative members based on a previously published sequence similarity network (SSN) analysis that partitioned the TSF proteins into five families; MIF, cis-CaaD, MSAD, CHMI, and 4-OT [1].
A multiple sequence alignment was performed utilizing the amino acid sequences of human MIF, human D-DT, Coryneform bacterium cis-CaaD, Pseudomonas aeruginosa CHMI, Pseudomonas pavonaceae MSAD, and Pseudomonas sp. (strain CF600) 4-OT (Figure S1). Considering the available crystal structures, deposited in protein data bank (PDB), our protein selection was made to ensure that the structural and dynamic properties of each protein family will be accurately analyzed. While the amino acid sequences come from different organism sources, the generated findings ensures broader applicability which is critical considering the size of this superfamily (>11,000). The highest sequence identity (SeqID) of 34.2% between any protein pair was obtained between MIF and D-DT (Table S1). The remaining protein pairs yielded values under 30%, with MSAD exhibiting similar SeqIDs with 4-OT (25.8%), cis-CaaD (25.0%), and CHMI (24.7%). Despite the low amino acid sequence identity, the six proteins demonstrate an overall satisfactory structural homology at the quaternary level, yet with diverse C-terminal segments (Figure 1A). Specifically, key differences were noted in the length (Figure S1) and secondary structure organization of the C-terminal tail (Figure 1A) as well as the position of the C-terminus in relation to the active site pocket (Figure 1B). For MIF, D-DT, MSAD, and 4-OT, the C-terminus is proximal to the active site opening, whereas in the cases of cis-CaaD and CHMI, the C-terminus is distal from it (Figure 1B). Interestingly, the active site opening of cis-CaaD is completely blocked by the β8/α3 loop, which is part of the C-terminal tail.
2.2. Root Mean Square Fluctuation (RMSF) Analysis Across the Target Proteins Demonstrates Diverse Dynamic Profiles
Upon identifying these structural differences in the C-terminal region, we performed MD simulations and analyzed the RMSF profiles of the six proteins (Figure 2 and Figure S2). 1 μs trajectories were considered suitable for investigating the C-terminal motions [37], while each calculation was repeated in triplicate. Globally, the six proteins demonstrated similar fluctuations with an average RMSF value of 0.71 ± 0.2 Å, 0.69 ± 0.3 Å, 0.88 ± 0.7 Å, 0.67 ± 0.5 Å, 0.79 ± 0.5 Å, and 0.87 ± 0.7 Å for MIF, D-DT, cis-CaaD, CHMI, MSAD, and 4-OT, respectively (Table S2, Figure S2). To confirm the accuracy of our approach, we compared our findings with previously published RMSF values obtained from 1 μs trajectories. In the absence of any data for the bacterial proteins, we considered only data derived from the two human proteins, MIF and D-DT. The previously published RMSF values of 0.90 Å [34] and 0.70 Å [16], for MIF and D-DT, respectively, are in agreement with the findings of this study (0.71 ± 0.2 Å (MIF) and 0.69 ± 0.3 Å (D-DT)). Having confirmed the accuracy of our calculation, we performed an in-depth analysis of the RMSF results focusing on the poorly studied bacterial proteins. For clarity, the secondary structure features of cis-CaaD, CHMI, MSAD monomers, and 4-OT dimer are provided (Figure S3).
The high similarity between the RMSF profiles of MIF and D-DT is apparent even upon a brief examination. However, each of the four bacterial proteins exhibits characteristic RMSF patterns that suggest unique intra- and inter-subunit communication pathways (Figure 2). Notably, for the two human proteins, the highest RMSF value of any region does not surpass 2 Å. Meanwhile, cis-CaaD, CHMI, MSAD, and 4-OT enclose one highly flexible region with an RMSF value far exceeding 2 Å. For CHMI, cis-CaaD, and 4-OT, this region is located in the C-terminal, whereas in MSAD, the β3/β4 loop is highly flexible. The reduced flexibility noted in the C-terminal residues of MSAD more closely resembles what is observed in MIF and D-DT (Figure 2). From a structural point of view, the high flexibility of cis-CaaD in the C-terminal tail, is of a great functional interest as it influences the mobility of β8/α3 loop, which in turn blocks the opening to the active site (Figure 1B).
Regions with fluctuation values greater than 1σ of the mean RMSF value were marked using the secondary structure features of each protein (Figure 2 and Figure S3). Our findings show that the human proteins have more regions with statistically significant RMSF values in comparison to the bacterial ones. This finding is not attributed to the enhanced flexibility of MIF and D-DT, but rather, it is explained by the presence of highly flexible regions within the four bacterial proteins. For this reason, we overlayed the six profiles and examined their fluctuation features region-by-region (Figure 2). The varying secondary structural features and length of the six proteins account for the differences seen in the overlayed RMSF illustration. Despite this, a similar fluctuation pattern with values above the average was noted in the α1/β2 loop (corresponding to the α1/β3 loop of CHMI). This loop is located in the active site cavity of all the proteins and is specifically adjacent to the catalytic residue P1 (Figure 2 and Figure S4). With reference to the better studied human proteins, MIF and D-DT, this loop includes residues that are important for catalysis and ligand binding [17,18,22,38]. Excluding MSAD, fluctuation above the mean value was also noted for residues found in the β4/α2 loop of MIF/D-DT and the corresponding β5/α2 and β1/α1 loops of cis-CaaD/CHMI and 4-OT, respectively. This loop, which is also located next to the catalytic residue P1, harbors an active site residue: I64 in MIF and D-DT, R11 in 4-OT, R71 in CHMI, R73 in cis-CaaD, and R75 in MSAD. In all proteins, fluctuation of the C-terminal region was found to be significant, surpassing 1σ of the mean RMSF value.
2.3. Correlation Plots Expose Communication Pathways with Mechanistic Interest
Correlation analyses of the Cα atoms were performed for the six proteins included in this study (Figure 3A–F). Similar to the RMSF analysis, MIF and D-DT (Figure 3A,B) were only used as controls to validate our data against previously published findings [16,34]. Once the reproducibility of our approach was confirmed, our attention shifted to the four bacterial proteins whose communication pathways are unknown (Figure 3C–F).
In all four proteins, the β1 strand is strongly correlated with the two adjacent strands of the monomeric β sheet, dimeric in the case of 4-OT. This finding is consistent with previously published observations of the two human proteins [16,34] and it reflects the fundamental role of the β sheet in correlated motions [39,40]. With cis-CaaD being a notable exception, we found another shared correlation between the first residue of β1 and the strand defining the solvent channel opening; β5 for MSAD, β7 for CHMI, and β2 of the adjacent monomer for 4-OT (Figure S3). This correlation is of great importance for the two human proteins as it was shown to modulate their catalytic activities via allosteric coupling [7,41].
With the objective to discover mechanistic insights into cis-CaaD, CHMI, MSAD, and 4-OT, we probed characteristic communications for each protein. In cis-CaaD, T34 stands out as a key residue situated on the flexible α1/β2 loop (Figure 4A). Our analysis showed that this residue is strongly correlated with multiple domains across the biological assembly of cis-CaaD (Figure 3C). Within the same subunit, T34 communicates with the β8/α3 loop, the α3 helix, and the C-terminal tail, which are all found proximal to this residue (Figure 4A). Noteworthy, β8/α3 is the loop blocking accessibility to the active site pocket (Figure 1B). T34 of monomer A also forms inter-subunit communications with the α1/β2 loops of monomers B and C, including their T34 residues which are found ∼34 Å apart (Figure 4A). In addition, T34 of monomer A is correlated with the β8/α3 loop and the α3 helix of monomer B as well as the three C-termini of cis-CaaD. These communications take place via long-range inter-subunit crosstalk with the participation of β5/α2 and β6/β7 loops as well as segments of the α2 helix and the β6 strand (Figure S3). With the exception of the β5/α2 loop, the remaining regions are located in the subunit-subunit interface and enable communications with the adjacent monomer. These findings led to the conclusion that catalysis in cis-CaaD is a highly coordinated process, where T34 plays a key role via major conformational changes in the C-terminal region β8/α3 loop, α3 helix, and the C-terminal tail.
For CHMI, an interesting observation is that all the C-termini are correlated with one another, despite being situated ~47 Å apart (Figure 3D). These correlations are enabled through the participation of multiple regions, including the α1/β3 loop that is strongly linked with the C-terminus (Figure 4B). Interestingly, the β8 strand that is found just before the C-terminus packs against the adjacent subunit. When examining subunit A, we observe that the β8 strand of this subunit interacts with the β7 strand of subunit C, bringing the C-terminus of subunit A proximal to the β6/β7 loop of subunit C (Figure 4B). The β6/β7 loop of subunit C is highly correlated with the β4/β5 loop of the same subunit, as well as the α1/β3 loop of subunit A. These correlated motions initiate from the highly flexible C-terminal region of subunit A, propagate through the β6/β7 loop of subunit C, and continue into the β4/β5 loop of the same subunit, ultimately reaching the α1/β3 loop of subunit A (Figure 4B). The α1/β3 loop of CHMI is characterized as the second highest RMSF region, surpassed only by the C-terminus. The correlated motions travel up towards the ends of β5 and β7 and, ultimately, reach the C-terminal solvent channel opening. Residues located at the end of β7 and within the β7/β8 loop of subunit A are correlated with the same residues of subunit C due to proximity, effectively bridging the C-terminus of subunit A and the C-terminus of subunit C.
From the RMSF analysis (Figure 2), it was clearly shown that MSAD exhibits distinct dynamic characteristics from cis-CaaD, CHMI, and 4-OT. A unique aspect of MSAD is that the region with the highest RMSF value is not the C-terminus, as is typically observed in the other three bacterial proteins, but rather the loop between β3 and β4 strands (Figure 4C). The correlation analysis also highlights that the β3/β4 loop is a region of high mechanistic value forming multiple strong intra- and inter-subunit communications across the biological assembly of MSAD (Figure 3E). The α1/β2 loop also exhibits a significant number of correlations, second only to the β3/β4 loop. The communication network formed through the β3/β4 loop as well as the α1/β2 loop facilitate the correlated motions within MSAD. Further analysis showed that the β3/β4 loop from different subunits form strong correlations with each other through intermediate communications that involve α1/β2 loop, α1, α2 helices, and α2/β5 loop (Figure 4C).
In the case of 4-OT, the communication pathways found in the biological assembly are more complicated in comparison to the other three proteins due to the homohexameric structure. For comparative analysis with the other TSF proteins, a dimer of 4-OT effectively acts as a pseudo-monomer, thereby incorporating two C-terminal regions within one monomer (Figure S3). Monomers A&D, B&E, and C&F form three homodimers, each of which corresponds to a monomer of cis-CaaD, CHMI, and MSAD. Analysis of the correlation data showed that the six C-termini communicated with each other despite the long distances (the shortest determined at ~26 Å). The flexibility observed in the C-terminal residues of the protein (Figure 2) plays a key role in the intra- and inter-subunit communication pathways enabling cross-talking between the C-termini. Our findings demonstrate that the communication pathway between the two C-termini of a given homodimer (pseudo-monomer) differs from the corresponding pathways formed across two homodimers (Figure 4D). Using the homodimer A/D as an example and in the order described, the C-terminus of monomer A communicates with the C-terminus of monomer D through the α1/β2, α1 and β1/α1 loops (Figure 4D). Communication between the C-terminus of monomer A and monomer C, which is a subunit of the adjacent homodimer, occur through the β3 strand, β2/β3 loop, β2 strand, and α1 helix (Figure 4D).
2.4. Correlation Analyses Reveals Communications Between the C-Terminal Region and Active Site Residues
As previously described, communications between the C-terminal region and active site residues modulate the enzymatic activities of MIF and D-DT. Bearing this in mind, we utilized our correlation analyses to detect distinct communication pathways in cis-CaaD, CHMI, MSAD, and 4-OT. Among the six proteins of this study, cis-CaaD is the largest, featuring a highly flexible C-terminal region (Figure 2) and exhibiting high intramolecular correlation with the α1 helix (Figure 3C). The active site residue H28 lies on this helix, but the correlated motions continue through the α1 helix towards R70 and R73 (Figure 5A), which are also active site residues.
For CHMI, our findings demonstrate cross-talk between residues situated at the interface of two monomers that bridge the C-terminal region with the catalytic residue P1 (Figure 5B). Specifically, the C-terminal region of subunit A is correlated to P1 of the same subunit via the β6/β7 loop derived from subunit C. From the β6/β7 loop, the correlated motions continue towards the β4/β5 loop of subunit C, then onto the α1/β3 loop of subunit A to eventually reach P1 (Figure 5B).
In MSAD, the C-terminal region exhibits greater restraint; however, a sizable and profoundly dynamic region of the β3/β4 loop within one subunit is positioned between the C-terminal segment and a loop adjacent to the active site residues of another monomer. This β3/β4 loop interfaces with a distinct dynamic α1/β2 loop of another monomer at the interphase between monomers. The C-terminal region of subunit A is highly correlated to the active site residue D37 via the highly fluctuating β3/β4 loop region of subunit C that lies between the two areas. The active site residue R73, located just after β4, is also highly correlated to the C-terminal region because of its proximity (Figure 5C).
4-OT stands out due to its deviation from being a homotrimer. For comparative analysis with the other TSF proteins, a dimer of 4-OT is regarded as a pseudo-monomer and thus, contains two C-terminal segments. Focusing on the catalytically relevant residues located on monomer A, we observe that the C-terminal region of monomer A at K59 demonstrates intra-subunit correlations to the active site residue F50, which is located at the beginning of the β3 strand (Figure 5D). Besides the intra-subunit correlations, the C-terminal region of a given monomer was found to form inter-subunit correlations with active site residues from adjacent monomers. For example, S58 derived from the C-terminal region of monomer C communicates with the active site residue R39 of monomer A, via P56 and I52 of monomer C (Figure 5D).
These correlation analyses have revealed robust communication between the C-terminal residues and crucial active site residues in every TSF protein included in this study. Through these correlated motions, the C-terminal region dynamically influences active site residues and strongly suggests that their impact in catalysis should be further explored with kinetic assays, utilizing C-terminal variants.
Source link
Christopher Argueta www.mdpi.com