1. Introduction
Rice (
Oryza sativa L.) is a fundamental staple food for approximately half of the global population, rendering it one of the most critical food crops worldwide [
1]. However, rice production faces significant challenges, particularly from rice blast disease, caused by the ascomycete fungus
Magnaporthe oryzae (syn.
Pyricularia oryzae) [
2,
3]. This disease is recognized as one of the most destructive fungal diseases in agriculture globally, posing a severe threat to rice cultivation. Statistics indicate that the global economic loss attributed to rice blast exceeds USD 7 billion annually [
4]. The rice yield reduction in fields where rice blast occurs is about 10% to 30% on average [
5], with severe outbreaks potentially leading to complete crop failure [
6]. The most cost-effective, efficient, and environmentally sustainable strategy for mitigating rice blast is to deploy resistance genes to develop resistant rice cultivars [
7,
8,
9].
Through long-term and extensive genetic analysis and research worldwide, nearly 118 rice blast resistance genes have been identified [
10], and among them, about 38 genes have been cloned [
11,
12,
13]. Genetic analysis has revealed that most of these resistance genes are dominant, while a few, such as
pi21 and
Pi55(t), are recessive [
14,
15]. Structural and functional analysis revealed that with the exception of
Pid2,
pi21, and
Ptr, which encode proteins with unique structures, most resistance genes encode proteins with coiled-coil (CC), nucleotide-binding site (NBS), and leucine-rich repeat (LRR) domains [
16,
17]. These resistance genes are unevenly distributed across 12 chromosomes of rice, with a concentration on chromosomes 6, 11, and 12, exhibiting a clustered distribution pattern [
18].
The accurate identification of resistance genes in rice germplasm is essential for their effective utilization in breeding programs. Traditionally, this has been accomplished by selecting specific pathogenic strains of
M. oryzae and inoculating rice cultivars to assess resistance or susceptibility, thus inferring the presence of particular resistance genes [
19]. However, this kind of work is time-consuming and inefficient, with results often being unreliable due to variability in rice growth conditions and inoculation processes [
20]. Compared with inoculation experiments, the application of molecular markers is more convenient and saves time. With the identification and fine mapping of resistance genes, a number of molecular markers closely linked to target resistance genes have been developed and applied in practice, such as molecular marker-assisted selection (MAS) breeding [
21]. Despite this, the accuracy of using closely linked markers for identifying target genes could be affected by linkage disequilibrium breakdown, population specificity, genetic recombination, as well as a lack of causal relationship with resistant phenotype. Additionally, closely linked markers often fail to differentiate between alleles at the same locus [
8,
22]. As rice blast resistance genes continue to be cloned, their complete gene sequences have become available, facilitating the development of gene-specific molecular markers. These markers enable the direct selection of target genes, overcoming the limitations associated with closely linked markers and ensuring greater selection accuracy [
23].
The
Pik locus, located at the terminus of the long arm of chromosome 11, comprises at least seven alleles:
Pik [
24],
Pi1 [
25],
Pikm [
26],
Piks [
27,
28],
Pikp [
29],
Pikh [
30], and
Pike [
31]. The resistance conferred by genes at the
Pik locus is governed by two adjacent CC-NBS-LRR genes (such as
Pik-1 and
Pik-2) with opposite transcriptional orientations [
26]. Studies targeting Pikm have found that the nucleic acid polymorphism is primarily associated with
Pikm-1, with variation concentrated in the CC domain, whereas
Pikm-2 remains more conserved [
32]. A mechanistic model of
Pikh-mediated resistance proposes that
Pikh-1 functions as an adaptor, facilitating the interaction between
AvrPik-h and
Pikh-2. Subsequently,
Pikh-2 transduces the signal to trigger
Pikh-specific resistance [
30]. The
Pik locus is critically important due to the application of its alleles in rice breeding programs aimed at achieving durable resistance against rice blast [
33]. The allele
Pik confers high and stable resistance to a wide range of Chinese rice blast isolates [
24]. Likewise, the
Pi1 allele originally derived from the West African cv. LAC23 [
25] has exhibited durable and high-level resistance to rice blast [
34]. Furthermore, Liu et al. [
35] reviewed the broad-spectrum resistance against
M. oryzae by
Pikh and
Pi1.
Yunnan is not only a center of diversity for Asia-cultivated rice (
O. sativa) but also the origin of rice blast fungus (
M. oryzae) [
36,
37], which presents unique opportunities for rice blast resistance breeding. To harness Yunnan’s rich genetic resources, it is imperative to accurately and swiftly identify resistance genes. At the same time, the
Pik locus offers vital materials for exploitation in resistance breeding. Currently, six alleles at the
Pik locus, excluding
Pike, have been cloned and published sequences. However, as new allele sequences are identified, previously developed markers may lose accuracy. In this study, the sequence information of these six alleles was analyzed in order to develop more precise and effective gene-specific molecular markers. These newly developed markers were subsequently applied to assess 163
japonica rice cultivars bred in Yunnan from 1980 to 2020, with the goal of determining the distribution of these six resistance genes at the
Pik locus. The aim of this research is to provide valuable insights and guidance for selecting resistance gene combinations in future rice breeding programs.
3. Discussion
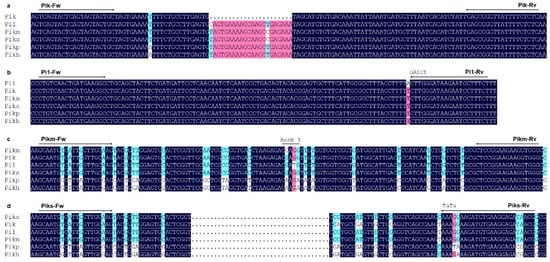
In this study, we successfully developed gene-specific molecular markers for six alleles at the Pik locus using publicly accessible gene sequences. These markers have been proven effective in accurately identifying target genes within rice genetic resources. Furthermore, a novel haplotype of Pik locus was found during the process of marker development. This new haplotype contains nucleotide mutations in its CDS region compared to known functional alleles, resulting in changes to the corresponding amino acid sequence, including a 9 bp insertion that encodes three additional amino acids. Based on these unique characteristics, we have developed haplotype-specific molecular markers to analyze the distribution of this new haplotype among improved rice cultivars of Yunnan.
The application of seven sets of specific markers developed by this study to detect 163 developed
japonica rice cultivars from the 1980s to 2020s in Yunnan revealed a limited utilization of
Pik locus resistance genes. Specifically, 38.65% of the cultivars, spanning all seven breeding series, carry the
Piks allele, which confers a low resistance frequency against
M. oryzae in the field. In contrast, the remaining five alleles of the
Pik locus are rarely utilized, with detection frequencies of 1.84% for
Pikm, 0.61% for
Pik, and 0% for
Pi1,
Pikp, and
Pikh, respectively. This underutilization of
Pik locus resistance genes restricts their potential application in rice breeding programs. Moreover, the long-term reliance on a single resistance gene can significantly increase selection pressure and accelerate the breakdown of resistance [
38]. Interestingly, 21.47% of the cultivars, distributed across all seven breeding series, do not carry any of the known
Pik locus alleles. This observation highlights the genetic diversity of Yunnan’s rice genetic resources and suggests the presence of unknown genes/alleles that may harbor new resistance functions. Despite the high sequence similarity among the coding sequences of
Pik locus alleles, their resistance functions differ significantly. Chen et al. [
31] tested the resistance spectra of these six alleles of
Pik locus with 215
M. oryzae isolates collected from different regions of China. The results showed a low resistance spectrum of
Piks (with a resistance frequency of 13.0%), medium resistance spectrum of
Pikp (31.6%),
Pik (39.1%),
Pi1 (48.4%),
Pikm (50.2%), and
Pikh (51.6%). However, 38.65% of the 163
japonica rice cultivars bred in Yunnan carry
Piks, while the other alleles were rarely utilized. This imbalance further underscores the limitations of current
Pik locus allele usage in rice breeding. The development of these sets of gene-specific markers will facilitate the rapid identification of
Pik locus alleles carried by rice genetic resources, expand the utilization of
Pik locus alleles, and facilitate the pyramiding of
Pik genes with other resistance genes to enhance both the resistance spectrum and the longevity of resistance in subsequent rice breeding efforts.
By integrating the latest sequence information, we have developed six sets of accurate and practical gene-specific markers for detecting alleles at the
Pik locus in Yunnan rice cultivars. Our findings revealed that alleles at the
Pik locus are limited in improved cultivars. This indicates that alleles such as
Pi1,
Pikm, and
Pikh, which exhibit good resistance against
M. oryzae [
39], could be strategically utilized in disease-resistant rice breeding in Yunnan to broaden the resistant spectrum. Notably, we identified a novel haplotype at the
Pik locus, with 37.42% of the improved Yunnan cultivars carrying this new haplotype. Further investigations into whether this haplotype represents a functional resistance allele are currently ongoing.