1. Introduction
The Chinese mitten crab,
Eriocheir sinensis (
E. sinensis), a member of the Grapsidae family of decapod crustaceans, is widely distributed in the East Asian–Pacific region and holds significant aquaculture value in China. Due to its high nutritional content and market demand,
E. sinensis is extensively cultured throughout China, with a production volume reaching 888,629 tons in 2023 [
1]. The cultivation of
E. sinensis supports a vital industry in eastern China, attracting increasing numbers of farmers. However, this expansion has introduced challenges such as sexual precocity [
2,
3], hepatopancreatic necrosis disease [
4], inbreeding, and the degeneration of genetic resources [
5]. These issues arise largely because many farmers rely on small-scale stock for breeding or indiscriminately hybridize geographically distinct wild populations with poor growth traits or lower taste quality in an attempt to reduce costs. Consequently, the genetic quality and overall performance of aquaculture stocks have deteriorated. To address these challenges, it is crucial to protect the genetic resources of
E. sinensis and guide the industry towards more sustainable breeding practices. Family-based selection, which has proven effective in the breeding of plants and livestock, holds great potential for aquaculture as well [
6]. By constructing large-scale families, breeders can accurately trace genealogical information, select high-quality parents, and avoid inbreeding. This approach also helps to continuously improve desirable traits and shortens the breeding cycle [
6,
7]. While family-based selection offers many benefits, it has not been widely adopted in
E. sinensis breeding due to the high costs associated with maintaining large family groups. Assigning each family to a specific pond can help minimize environmental variations and reduce the costs of maintaining separate family lines [
8]. Traditionally, physical tags have been used to distinguish individual lineages [
9,
10]. However, applying physical tags to
E. sinensis is problematic, as the crabs molt frequently, making it difficult to attach and maintain tags on their bodies. Implanting electronic tags may offer a solution, but the crabs need to reach a sufficient size, and the technology for reliable tagging is still under development. Tags are prone to falling off, and the mortality rate may increase if the tags are improperly applied. A more effective solution is to house different families in the same pond and use DNA molecular markers to distinguish between them.
Molecular markers such as random amplification polymorphism DNA (RAPD), amplified fragment length polymorphism (AFLP), microsatellites (also known as simple sequence repeat, SSR), and single-nucleotide polymorphisms (SNPs) are widely used in breeding programs [
11,
12]. While SNPs have been increasingly applied in genetic studies of crustaceans such as Pacific white shrimp (
Litopenaeus vannamei) [
13,
14], mud crab (
Scylla paramamosain) [
15], and swimming crab (
Portunus trituberculatus) [
16], their development and analysis require more advanced and costly technologies. In contrast, SSR markers remain a preferred tool for genetic diversity analysis [
17,
18] and parentage identification [
19,
20] due to their accessibility. Microsatellite markers have been widely utilized for over a decade due to their high abundance, variability, co-dominant inheritance, and neutral selection within the genome. They typically consist of tandem repeating units of mono-, di-, tri-, and tetra-nucleotide sequences [
21,
22]. Their high allelic diversity provides superior power for determining pedigree relationships among individuals [
23]. In
E. sinensis, microsatellite markers are commonly used for constructing linkage maps [
24,
25], assessing population genetic diversity [
26,
27,
28], and conserving and exploiting wild crab genetic resources [
29,
30]. For family management, the use of microsatellite markers in
E. sinensis offers several advantages, including reduced maintenance costs, savings in manpower and space, and a more reliable method for evaluating genetic parameters. However, despite these benefits, the cost of genotyping large numbers of individuals with microsatellite markers can still be considerable.
Establishing microsatellite multiplex PCR panels is useful for evaluating parentage assignment in applied fisheries, particularly when managing large numbers of families. Multiplex PCR panels reduce the time and cost associated with genetic diversity evaluations using SSR markers, minimizing repetitive manipulations and manual handling errors [
31]. These panels have been successfully implemented in several aquatic species, including grass carp (
Ctenopharyngodon idellus) [
32], mussel
Anodonta (
Sinanodonta)
woodiana (Lea, 1834) [
33], and swimming crab (
Portunus trituberculatus) [
34]. However, research on the development of multiplex PCR systems for
E. sinensis remains limited, despite their potential to enhance large-scale family breeding programs.
With the advent of next-generation sequencing technology for transcriptomics, the acquisition of a substantial number of SSR markers has become more cost-effective. In this study, we sequenced the transcriptomics of E. sinensis, identifying 1,521,519 putative microsatellite markers. From 272 designed SSR primer pairs, 205 were validated, with 100 demonstrating high polymorphism. By screening these polymorphic microsatellite markers, we developed four microsatellite multiplex PCR panels for the first time in E. sinensis and applied them to determine parentage in six families. This approach significantly enhances analytical efficiency and offers a practical and effective method for breeding and family management of E. sinensis, promoting the sustainable development of the industry.
4. Discussion
With the advent of next-generation sequencing technology, the discovery of SSR markers is no longer confined to constructing SSR libraries and sequencing candidate clones. Given the challenges of obtaining high-quality genome-wide information for non-model organisms, an increasing number of studies have shown that high-throughput microsatellite marker development from transcriptome data is an effective approach [
45]. In this study, we conducted transcriptome sequencing of the ovaries of
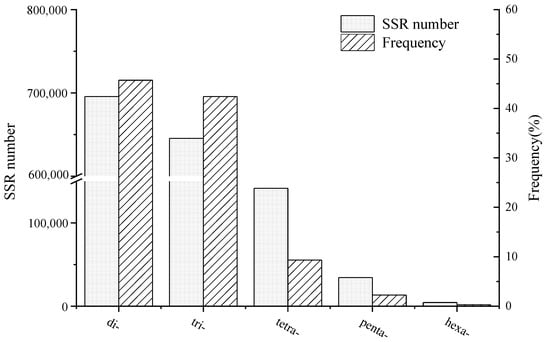
Eriocheir sinensis at different developmental stages using the Illumina HiSeq™ 2500 platform, resulting in the identification of 1,521,519 candidate microsatellite markers. Among these SSR markers, di-nucleotide repeat motifs were the most prevalent (45.72%), followed closely by tri-nucleotide repeat motifs (42.42%). These findings align with previous reports on
E. sinensis, which indicated that di-nucleotide repeat motifs were the most abundant type (58.54%), followed by tri-nucleotide repeats (30.11%) [
28]. It is generally recognized that most SSR repeats in animals are di-nucleotide repeats [
46,
47], and our results support this observation. To validate the identified SSR markers, we randomly selected a subset of 272 primer pairs for PCR, achieving a high polymorphism rate of 48.8% (100 out of 205) for the SSR markers. The lower polymorphism rates observed in our study compared to previous reports on
E. sinensis may stem from false positives inherent in transcriptome sequencing, as well as variations in SSR search tools and criteria employed [
48]. The number of SSR markers found in the transcriptome was lower, which is expected since transcriptome sequences primarily represent functional genes with fewer repeat sequences in coding regions; most repeated sequences are typically found in the untranslated regions (UTRs). Additionally, the challenge of amplifying larger intron regions can hinder the efficiency of microsatellite development in the transcriptome, necessitating the selection of primers that amplify smaller fragments. However, microsatellite markers developed from the transcriptome are often derived from regulatory or coding regions, enabling the direct and accurate tagging of functional genes, which is beneficial for studying functional variation, adaptive evolution, and genetic diversity in species during genetic processes [
49,
50]. In summary, this transcriptome provides a valuable resource for future gene analyses, and these microsatellite markers will be instrumental in advancing our understanding of the molecular ecology and genetic breeding of
E. sinensis.
The establishment of microsatellite multiplex PCR represents the most efficient method for evaluating parentage assignment in applied fisheries, especially when a large number of family separations is required. This study is the first to establish a microsatellite multiplex PCR panel for
E. sinensis. We developed four multiplex panels, each containing four microsatellite markers, designed based on allelic size range and compatibility of fluorescent labeling dyes. A critical factor in establishing and applying a multiplex microsatellite assay is the appropriate selection of sites and their combinations [
51]. The concentration of each primer in the multiplex PCR reaction system is particularly crucial, albeit a tedious and challenging step. Various factors influence primer concentrations, including annealing temperature, nucleotide sequence patterns, and PCR product size [
34,
52]. Research suggests that the size difference between fragments at two markers should exceed 20 bp to minimize potential biases in amplification [
53]. In our assay, the product fragment sizes between markers were greater than 20 bp or labeled with different fluorescent markers, allowing for accurate allele peak distinction. Ultimately, we successfully developed four SSR multiplex PCR sets for
E. sinensis, which can serve as effective tools for genetic studies, including population genetic structure assessments and parentage assignments.
Beyond
E.
sinensis, multiplex PCR has been widely applied in other aquatic species. For instance, in the giant freshwater prawn (
Macrobrachium rosenbergii), multiplex PCR has been utilized for genetic analysis and parentage verification. Similarly to our findings, studies on
Macrobrachium rosenbergii have emphasized the efficiency of multiplex PCR in reducing time and reagent costs while maintaining high accuracy in genetic assessments. However, it is noteworthy that genetic diversity in
Macrobrachium rosenbergii populations tends to be lower than that in
E. sinensis [
54,
55]. In grass carp (
Ctenopharyngodon idellus), multiplex PCR has been successfully applied to evaluate genetic diversity in breeding programs, revealing higher genetic diversity in cultured populations compared to
E. sinensis, with a mean Na of 22.15, mean Ho of 0.823, mean He of 0.859, and mean PIC of 0.846 across nine populations [
32]. The successful application of multiplex PCR across various aquatic animals underscores its value as a versatile and powerful tool for genetic analysis and breeding management.
Microsatellite markers have been extensively used for parentage assignment in various aquatic animals, such as orange-spotted grouper (
Epinephelus coioides) [
56], tiger trout (
Salmo trutta ×
Salvelinus fontinalis) [
57], and red swamp crayfish (
Procambarus clarkii) [
58]. These markers have significantly reduced the workload associated with distinguishing different families compared to traditional physical tagging methods. In our study, parentage assignment was performed on six families using microsatellite multiplex PCR, achieving 100% accuracy in assigning offspring to their parents when three or four multiplex PCR sets were utilized. Varying success rates for parentage assignments have been reported previously; for example, Zhang et al. (2016) used nine SSR markers to identify 18 full-sib families in yellow catfish
Pelteobagrus Fulvidraco, achieving 100% allocation for all progeny [
59]. Similarly, Satyanarayana et al. (2015) achieved a 91% assignment success rate using four SSR markers in freshwater prawn
Macrobrachium rosenbergii [
60]. Yang et al. (2014) reported a 97% success rate with ten SSR markers in mandarin fish
Siniperca chuatsi [
61]. The success rate observed in our study meets the practical needs for production applications.
Our findings suggest that the accuracy of parentage assignment depends on the number of markers used and their polymorphism; a lower-locus polymorphism necessitates a higher number of markers for reliable assignment. Previous studies have emphasized that selecting microsatellite markers with high polymorphism enhances the power of parentage assignment [
59,
62]. Moreover, the ability to identify paternity diminishes as the number of candidate parents increases, underscoring the need for an increased number of molecular markers alongside the growing number of parents and offspring. For instance, it was shown that with the same number of microsatellite markers, identification rates were 92.2% with three pairs of candidate parents but only 78.9% with eight pairs [
63]. While higher identification rates can be achieved with a greater number of microsatellite markers, this approach also increases workload and optimal number of markers. In our study, 100% of the offspring were accurately allocated to their parents when using 12 microsatellite markers.
Additionally, our results indicate that the simulated assignment success rates were higher than the actual assignment rates. This discrepancy may be attributed to genotyping errors and the presence of null alleles, which are known to impact the accuracy of parentage assignments [
64]. Such issues could arise from mutations or indels at one or both of the primer-binding sites [
65]. Similar phenomena have been reported in studies involving the pacific oyster
Crassostrea gigas [
66] and yellow catfish [
59]. Notably, some markers (
CX140,
CX416,
CX003) exhibited high null allele frequencies; however, the error rates in parentage assignments can be mitigated by utilizing a combination of all markers, enabling the correct identification of offspring and assignment to their most likely parent.