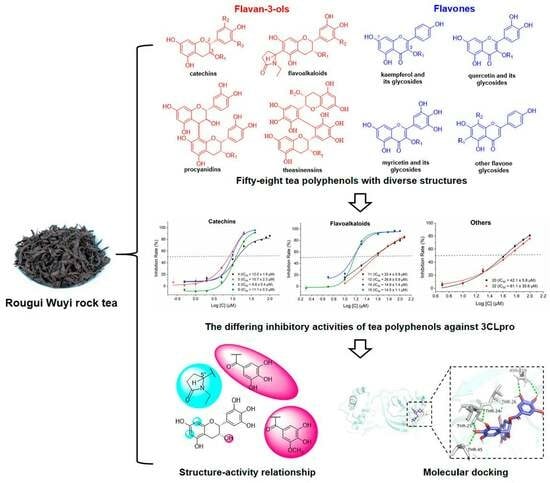
2.1. Isolation and LC-MS Analysis of RGWRT
In order to obtain the diverse structures of tea polyphenols, the chemical components of RGWRT were systematically studied. As a result, fifty-eight polyphenolic compounds were afforded and could be classified into two main categories: flavan-3-ols (
1–
21) and flavones (
22–
58). These isolated substances were used as standards in the UPLC-Q-TOF-MS analysis of RGWRT extract. The chromatographic peaks were identified by the comparison with retention time, accurate parent mass, and fragment ion spectrogram of standards, and the results confirmed that the main tea polyphenols have been obtained in the present study (
Figure 1). Furthermore, the flavan-3-ols in RGWRT could be further subdivided into several subgroups, including catechins, flavoalkaloids, procyanidins, theasinensins and their corresponding derivatives. The flavones could be further categorized into different classes based on the number of hydroxyl groups, encompassing kaempferol, quercetin, and myricetin, along with their respective
O-glycosides. The
C-glycosides of flavone are presented as a distinct subclass. The specific structures of various types of tea polyphenols are illustrated in
Figure 2. This is the first time that a wide variety of tea polyphenols with diverse structural types were obtained for sequential studies.
The structures of all polyphenols were elucidated on the basis of comprehensive analysis of 1D and 2D NMR and HRMS spectroscopic data, including eleven undescribed compounds (
11,
12,
20,
30,
32,
34,
36,
43,
44,
46, and
48) and forty-seven known tea polyphenols. The known compounds were identified by comparing their spectral data with those reported in the literature as (−)-epicatechin (
1) [
34], (−)-epicatechin-3-
O-gallate (
2) [
35], (−)-epicatechin-3-(3′-
O-methyl)-gallate (
3) [
36], (−)-epigallocatechin (
4) [
37], (−)-epigallocatechin-3-
O-gallate (
5) [
38], (−)-epigallocatechin-3-(3′-
O-methyl)-gallate (
6) [
39], (−)-epiafzelechin-3-
O-gallate (
7) [
40], (+)-catechin (
8) [
41], (−)-gallocatechin-3-
O-gallate (
9) [
38], etc-pyrrolidinone F (
10) [
42], puerin IV (
13) [
19], puerin V (
14) [
19], puerin VI (
15) [
19], procyanidin B2 (
16) [
43], procyanidin B2-3′-
O-gallate (
17) [
44], procyanidin B4-3′-
O-gallate (
18) [
45], catechin-(4
α→8)-epigallocatechin-3-
O-gallate (
19) [
46], theasinensin A (
21) [
47], kaempferol (
22) [
48], astragalin (
23) [
49], kaempferol-3-
O–
β-D-galactopyranoside (
24) [
50], kaempferol-3-
O-rutinoside (
25) [
51], kaempferol-3-
O-robinobioside (
26) [
52], kaempferol-3-
O-[
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucopyranoside (
27) [
53], kaempferol-3-
O-[
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-galactopyranoside (
28) [
54], kaempferol-3-
O-[2″-
O-(
E)-
p-coumaroyl] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucopyranoside (
29) [
55], kamoreokchaside I (
31) [
56], camellikaempferoside A (
33) [
57], camellikaempferoside C (
35) [
58], quercetin-3-
O–
β-D-glucoside (
37) [
59], quercetin-3-
O–
β-D-galactoside (
38) [
59], rutin (
39) [
60], quercetin-3-
O-[
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucopyranoside (
40) [
53], quercetin-3-
O-[
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-galcopyranoside (
41) [
53], camelliquercetiside B (
42) [
61], camelliquercetiside C (
45) [
61], camelliquercetiside A (
47) [
61], myricetin-3-
O–
β-D-glucopyranoside (
49) [
62], myricetin-3-
O–
β-D-rutinoside (
50) [
63], isovitexin (
51) [
64], isovitexin-2″-
O-glucoside (
52) [
65], apigenin-6-
C–
α-L-rhamnopyranosyl-(1→2)-
β-D-glucopyranoside (
53) [
66], apigenin-6-
C–
α-L-rhamnopyranosyl-(1→2)-
α-L-arabinopyranoside (
54) [
67], schaftoside (
55) [
68], isoschaftoside (
56) [
53], vitexin (
57) [
69], vitexin-2″-
O-glucoside (
58) [
65], respectively.
2.2. Structure Elucidation of New Tea Polyphenols from RGWRT
Compound
11 was obtained as white amorphous powder. The molecular formula was deduced as C
21H
23NO
8 from a molecular ion peak at
m/z 416.1331 [M − H]
− (calcd for C
21H
22NO
8, 416.1345) in the HR-ESI-MS. The presence of a flavan-3-ol skeleton was inferred obviously from the
1H and
13C NMR spectra (recorded in methanol-
d4) of
11 (
Table 1,
Figure S2) [
37]. More specifically, the typical proton signals at
δH 6.01 (1H, s) for ring A, at
δH 6.51 (2H, s, H-2′, H-6′) for ring B, and at
δH 4.76 (1H, s, H-2),
δH 4.20 (1H, s, H-3),
δH 2.86 (1H, m, H-4
β),
δH 2.76 (1H, m, H-4
α) for ring C, were similar to those of (−)-epigallocatechin (EGC) (
Table 1). A single proton signal on the A ring indicated a replacement at C-6 or C-8. Besides the similar signals with EGC skeleton, the
1H NMR spectrum of
11 indicated the presence of three sets of methylene signals at
δH 2.66, 2.42 (each 1H, m) for H-3″,
δH 2.17 and 2.34 (each 1H, m) for H-4″,
δH 2.66 and 3.50 (each 1H, m) for H-6″, a methine proton at
δH 5.45 (1H, s) for H-5″, and one methyl group at
δH 1.01 (3H, t,
J = 7.2 Hz) for H-7″, and
13C NMR spectrum showed the corresponding signals attributable to one carbonyl (
δC 177.6, C-2″), two methylenes (
δC 32.6, 24.5, C-3″ and 4″), a methine (
δC 54.1, C-5″), and an ethyl group (
δC 36.3, 12.6, C-6″ and 7″), which was further supported by HSQC correlations. The HMBC correlations between H-3″ with C-5″, C-4″, and C-2″ suggested the presence of a
N-ethyl-2-pyrrolidinone ring [
70] (
Figure 3). The existence of a partial structure of –C(3″)H
2−C(4″)H
2−C(5″)H− could be further inferred through the
1H−
1H COSY correlations of H-4″ with H-3″/H-5″ [
42]. The combination of the HMBC correlations between H-4″ to C-6/C-2″/C-5″/C-3″ and H-8 to C-6/C-7/C-9/C-10, as well as the ROESY (recorded in DMSO-
d6) correlation of the signal (
δH 5.89, H-8) with the 7-OH proton (
δH 9.23) (
Figure S8) showed that the
N-ethyl-2-pyrrolidinone moiety was connected to the C-6 position [
71]. Thus, the planar structure of compound
11 was determined as 6-
N-ethyl-2-pyrrolidinone-epigallocatechin.
Compound
12 was found to have the same molecular formula as
11 (C
21H
23NO
8) based on a molecular ion peak at
m/z 416.1336 [M − H]
− (calcd for C
21H
22NO
8, 416.1345) in the HR-ESI-MS. The
1H NMR and
13C NMR spectra of
12 were similar to those of
11, indicating they possess the same structural scaffold. The position of
N-ethyl-2-pyrrolidinone in compound
12 was also determined to be at C-6 via the similar key correlations of ROESY (recorded in DMSO-
d6,
Figure S17) and HMBC spectra (recorded in methanol-
d4,
Figure S16).
The difference between
12 and
11 is the configuration of C-5″, and previous studies have shown that the absolute configuration at C-5″ can be distinguished by subtracting one CD spectrum from its stereoisomer with the same configurations at C-2/C-3 [
19,
42]. To identify the absolute configuration at C-5″ of compounds
11 and
12, a pair of stereoisomers at C-5″, the CD spectra of two compounds were compared after subtracting the CD spectrum from each other (
Figure 4) [
42]. For compound
11, the arithmetically independent CD curves of C-5″ exhibited a strong negative cotton effect at 211 nm (Δε − 14.0) and was confirmed to be of the C-5″
R configuration. The configuration of
12 was deduced as
S configuration due to a positive cotton effect at 211 nm (Δε + 14.0) in the CD spectrum (
Figure 4). Therefore, the structures of compound
11 and
12 were determined to be 6-(5″
R)-
N-ethyl-2-pyrrolidinone-epigallocatechin and 6-(5″
S)-
N-ethyl-2-pyrrolidinone-epigallocatechin.
Compound
20 was determined to have a molecular formula of C
38H
32O
18 based on HR-ESI-MS at
m/z 775.1505 [M − H]
− (calcd for C
38H
31O
18, 775.1510). The
1H and
13C NMR data of
20 were similar to theasinensin B except for the addition of a methoxy group [
δH 3.81, (3H, s)], indicating that
20 was a methylated theasinensin B [
45]. The HMBC correlations from the methoxy proton to C-3″, and from H-2″ (
δH 6.99) to C-1″/C-3″/C-4″/C-6″/C-7″ (
Figure 3) confirmed that the methoxy group was linked to C-3″. Therefore, the structure of compound
20 was assigned as epigallocatechin-(2′→2′)-epigallocatechin-3-(3″-
O-methyl)-gallate.
Compound
30 was presented as yellow amorphous powder with a molecular formula of C
42H
46O
22 by HR-ESI-MS ion at
m/z 901.2394 [M − H]
− (calcd for C
42H
45O
22, 901.2402). The
1H NMR and
1H−
1H COSY spectrum of
30 suggested the presence of characteristic kaempferol aglycone [
δH 6.19, (1H, d,
J = 2.0 Hz) and 6.38 (1H, d,
J = 2.0 Hz) for A ring;
δH 6.91 (2H, d,
J = 8.7 Hz) and 8.00 (2H, d,
J = 8.7 Hz) for B ring],
Z–
p-coumaroyl group [
δH 6.70 (2H, d,
J = 8.5 Hz), 7.64 (2H, d,
J = 8.6 Hz), 5.89 (1H, d,
J = 12.7 Hz) and 6.88 (1H, d,
J = 12.7 Hz)] [
72], and three monocarbohydrate moieties [
δH 4.41 (1H, d,
J = 7.7 Hz), 4.55 (1H, br s) and 5.51 (1H, d,
J = 8.0 Hz) for three anomeric protons] (
Table 2). In addition, the corresponding anomeric carbons of the sugars were observed at
δC 105.7, 102.1 and 100.7 in the
13C NMR spectrum, with the help of the HSQC experiment. Acid hydrolysis of
30 followed by UPLC-CAD analysis identified two D-glucose, and a L-rhamnose. The key HMBC correlations of H-1″ (
δH 5.51, Glc) to C-3 (
δC 134.6), H-1‴ (
δH 4.55, Rha) to C-6″ (
δC 68.2, Glc), and H-1′′′′ (
δH 4.41, Glc) to C-3‴ (
δC 83.2, Rha) determined the linkage of sugar chain [
55,
56]. The
p-coumaroyl group was attached at C-2″ by the correlation between
δH 5.00 (H-2″) to
δC 167.6 (C=O of
Z–
p-coumaroyl group). Therefore, the chemical structure of
30 was determined to be kaempferol-3-
O-[2″-
O-(
Z)–
p-coumaroyl] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucopyranoside.
Compound 32, a yellow amorphous powder, showed an anion peak at m/z 901.2384 [M − H]− (C42H45O22, 901.2402) in HR-ESI-MS spectrum, exhibited similar 1H and 13C NMR spectra compared to 30. The main difference was that the glucopyranoside attached at C-3 in 30 was replaced by a galactopyranoside moiety, which was confirmed by the acid hydrolysis and the anomeric proton [δH 5.41 (1H, d, J = 7.9 Hz)]. Thus, compound 32 was elucidated as kaempferol-3-O-[2″-O-(Z)-p-coumaroyl] [β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→6)]-β-D-galactopyranoside.
Compound
34, a yellow amorphous powder, was determined as C
38H
48O
24 by HR-ESI-MS at
m/z 887.2448 [M − H]
− (calcd for C
38H
47O
24, 887.2457). The
1H and
13C NMR data of
34 were similar to those of
30 except for the addition of a sugar unit at C-3″ and the absence of
p-coumaroyl group, indicating that
34 was a kaempferol tetraglycoside. The presence of two
β-D-glucopyranosyl, one
α-L-arabinopyranosyl, and one
α-L-rhamnopyranosyl units were confirmed by the acid hydrolysis and coupling constants. The glycosidic linkage of oligosaccharide chain was established via the key HMBC correlations from
δH 5.17 (H-1″, Glc) to
δC 135.5 (C-3),
δH 4.56 (H-1‴, Rha) to
δC 68.8 (C-6″, Glc),
δH 4.40 (H-1′′′′, Glc) to
δC 83.2 (C-3‴, Rha), and
δH 4.57 (H-1′′′′′, Ara) to
δC 86.9 (C-3″, Glc) (
Figure 3). Thus, the structure of
34 was determined to be kaempferol-3-
O-[
α-L-arabinopyranosyl-(1→3)] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucopyranoside.
Compound 36 was purified as a yellow amorphous powder, and its molecular formula was deduced as C47H54O26 by HR-ESI-MS at m/z 1033.2805 [M − H]− (calcd for C47H53O26, 1033.2825). The NMR data obtained for 36 resembled those of 30, except for the additional arabinopyranosyl unit attached at C-3″, which was supported by the acid hydrolysis and HMBC correlation of H-1′′′′′ (δH 4.30) to C-3″ (δC 80.5). The α-configuration of the arabinopyranosyl group was determined by the coupling constant (J = 5.9 Hz) of the anomeric proton. Consequently, compound 36 was elucidated as kaempferol-3-O-[2″-O-(Z)-p-coumaroyl] [α-L-arabinopyranosyl-(1→3)] [β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→6)]-β-D-glucopyranoside.
Compound
43 was presented as yellow amorphous powder with a molecular formula of C
42H
46O
23 by HR-ESI-MS ion at
m/z 917.2331 [M − H]
− (calcd for C
42H
45O
23, 917.2352). The
1H NMR spectrum indicated the typical quercetin unit signals at
δH 7.46 (1H, d,
J = 2.2 Hz, H-2′),
δH 7.63 (1H, dd,
J = 8.5, 2.3 Hz, H-6′),
δH 6.79 (1H, m, H-5′),
δH 6.29 (1H, br s, H-8) and
δH 6.11 (1H, br s, H-6), and
E–
p-coumaroyl group with protons at
δH 7.56 (1H, d,
J = 15.9 Hz, H-Cou-7),
δH 7.52 (2H, d,
J = 8.6 Hz, H-Cou-2, H-Cou-6),
δH 6.79 (2H, d,
J = 8.4 Hz, H-Cou-3, H-Cou-5), and
δH 6.38 (1H, d,
J = 15.9 Hz, H-Cou-8) [
72]. Furthermore, three anomeric proton signals of sugar moieties were observed at
δH 5.59 (1H, d,
J = 8.0 Hz, H-1″),
δH 4.49 (1H, br s, H-1‴), and
δH 4.32 (1H, d,
J = 7.7 Hz, H-1′′′′) (
Table 2). 2D NMR and acid hydrolysis experiments were performed to determine the sugar unit and the glycosidic linkage of
43, which suggested the existence of a
β-D-galactopyranosyl, an
α-L-rhamnopyranosyl, and
β-D-glucopyranosyl moieties. The attachments of sugar chains and
E–
p-coumaroyl group were determined by the HMBC correlations of
δH 5.59 (H-1″, Gal) to
δC 132.8 (C-3),
δH 4.49 (H-1‴, Rha) to
δC 64.9 (C-6″, Gal),
δH 4.32 (H-1′′′′, Glc) to
δC 81.7 (C-3‴, Rha) and
δH 5.22 (H-2″, Gal) to δ
C 166.0 (C=O of
E–
p-coumaroyl group) [
61]. Accordingly, compound
43 was identified as quercetin-3-
O-[2″-
O-(
E)-
p-coumaroyl] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-galcopyranoside.
Compound
44 was obtained as a yellow amorphous powder, and its molecular formula was deduced as C
42H
46O
23 by HR-ESI-MS at
m/z 917.2332 [M − H]
− (calcd forC
42H
45O
23, 917.2352). The
1H and
13C NMR data of
44 exhibited high similarity to that of
43, with the main distinction being a configuration of coumaroyl moiety shift from
E– in
43 to
Z– in
44 (
Table 3), which was supported by the two olefinic protons at
δH 6.82 (1H, d,
J = 12.0 Hz) and
δH 5.81 (1H, d,
J = 12.9 Hz). Hence, the structure of compound
44 was established as quercetin-3-
O-[2″-
O-(
Z)-
p-coumaroyl] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-galcopyranoside.
Compound 46 was isolated as a yellow amorphous powder with a molecular formula of C38H48O25 by HR-ESI-MS ion at m/z 903.2400 [M − H]− (calcd for C38H47O25, 903.2406). The 1H and 13C NMR spectroscopic data of 46 were similar to those of 44 except for the addition of an arabinopyranosyl unit at C-3″ and the absence of Z–p-coumaroyl group, indicating that 46 was a quercetin tetraglycoside, which was further confirmed by the acid hydrolysis. The attachment of an arabinopyranosyl group to C-3″ was established from the HMBC correlation between H-3″ (δH 3.60, Glc) with δC 105.6 (H-1′′′′′, Ara). Consequently, compound 46 was elucidated as quercetin-3-O-[α-L-arabinopyranosyl-(1→3)] [β-D-glucopyranosyl-(1→3)-α-L-rhamnopyranosyl-(1→6)]-β-D-glucopyranoside.
Compound
48, a yellow amorphous powder, was determined as C
47H
54O
27 by HR-ESI-MS at
m/z 1049.2751 [M − H]
− (calcd for C
47H
53O
27, 1049.2774). Compound
48 showed similar NMR data to that of camelliquercetiside A [
62], except for slight differences of the two olefinic proton signals at
δH 6.82 (1H, d,
J = 13.3 Hz) and
δH 5.83 (1H, d,
J = 12.8 Hz), indicating the different configuration of the double bond in
48. Moreover, the linkage of the
Z–
p-coumaroyl unit was assigned at C-2″, which was confirmed through the HMBC correlation from H-2″ (
δH 5.04, m) to C-Cou 9 (the carbonyl carbon of the
Z-coumaroyl moiety,
δC 164.9). Therefore, the structure of compound
48 was characterized as quercetin-3-
O-[2″-
O-(
Z)-
p-coumaroyl] [
α-L-arabinopyranosyl-(1→3)] [
β-D-glucopyranosyl-(1→3)-
α-L-rhamnopyranosyl-(1→6)]-
β-D-glucoside.