1. Introduction
Myofibrillar protein (MP) constitutes the predominant fraction of fish muscle protein, accounting for approximately 50–55% of the total [
1]. It plays a crucial role in determining the structural integrity and functional properties of muscle proteins, including water retention [
2] and gel characteristics [
3], thereby influencing the texture and flavor of fish. The main constituents of MP, such as myosin heavy chain/light chain and actin, are highly abundant in fish muscle and particularly susceptible to oxidation [
4,
5,
6,
7]. Research indicates that oxidation of MP alters the physicochemical characteristics [
8], thereby significantly impacting the quality and nutritional value of fish muscle [
9,
10].
Fish, as a vital source of high-quality protein, provide essential amino acids, micronutrients, and polyunsaturated fatty acids, contributing to the daily nutritional needs of humans [
11]. However, during storage and processing, the presence of pro-oxidants, like unsaturated fatty acids and transition metal ions, renders MP highly vulnerable to oxidation [
12,
13]. This oxidative degradation leads to the loss of essential nutrients and causes undesirable changes in texture, WHC, color, and flavor while also promoting the formation of toxic substances. Given the pivotal role of MP in determining fish muscle quality, understanding the mechanisms and effects of MP oxidation is essential for mitigating quality deterioration and enhancing the storage, processing, and nutritional attributes of fish products.
Unsaturated fatty acids (UFAs) play an important role in the nutrition and flavor of foods [
14,
15]. UFAs are susceptible to oxidation, generating free radicals and reactive oxygen species (ROS) [
16,
17], which attack the amino acid residues on protein molecules, further inducing protein oxidation, resulting in structural and functional changes in proteins [
18,
19]. In grass carp, linoleic acid (OLA) serves as the primary source of polyunsaturated fatty acids [
20]. As a widely used UFA, OLA has also been utilized to construct lipid-induced protein oxidation models. Jiang et al. [
21] investigated the effect of OLA addition on the water holding capacity (WHC) of beef MP gel and indicated that a moderate concentration of OLA (≤6 mM) could increase the gel WHC, while a high concentration of OLA (>10 mM) led to protein aggregation, thereby reducing the gel WHC. Liao et al. [
22] explored the effects on the structure and rheological properties of carp MP gel under the addition of OLA and showed that the OLA moderate oxidative modification (≤2.5 mM) enhanced the gel-forming ability of myofibrillar protein (MP). OLA with a concentration below 3 mM markedly increased the digestion rate and digestibility of MP, while at high concentrations (5–10 mM), OLA induced the adverse effect in protein digestion [
23].
Current studies on MP oxidation induced by OLA are mostly conducted under ambient temperature conditions. However, fish are usually subjected to heat treatment before consumption, which inhibits the growth of microorganisms [
24], enhances the organoleptic quality [
25], as well as induces protein denaturation, facilitating digestion and absorption [
26]. Despite these benefits, heat treatment also leads to protein oxidation, which can negatively impact the fish meat quality and nutritional value [
27]. Yu et al. [
28] used a simulated digestion model to study the impact of oxidation on protein digestion in cooked abalone muscle. The results indicated that the boiling treatment caused protein oxidation in abalone muscle, significantly inhibiting the degree of hydrolysis and digestibility of the proteins. Zhang et al. [
29] extracted oyster salt-soluble proteins (OSSP) from Pacific oysters and examined the effects of heat treatment (35–100 °C) on the physicochemical properties and in vitro digestion of OSSP. The findings revealed that heating significantly reduced protein solubility and increased surface hydrophobicity and carbonyl content. In vitro digestion results demonstrated that protein oxidation and aggregation inhibited the digestion of OSSP [
29]. Additionally, Xia et al. [
30] investigated the effects of different roasting temperatures (150 °C, 190 °C, 230 °C, 270 °C, 310 °C) on protein oxidation and amino acid modifications in beef patties. The results showed that, as the roasting temperature increased, the level of protein oxidation intensified, amino acid side chains were modified, high temperatures significantly increased cooking loss, high temperatures elevated pH values, and high temperatures resulted in different color values. During heat treatment, the interaction between proteins and lipids can significantly influence protein oxidation. However, the impact of heat treatment on protein oxidation, specifically focusing on OLA-induced MP oxidation, remains insufficiently studied.
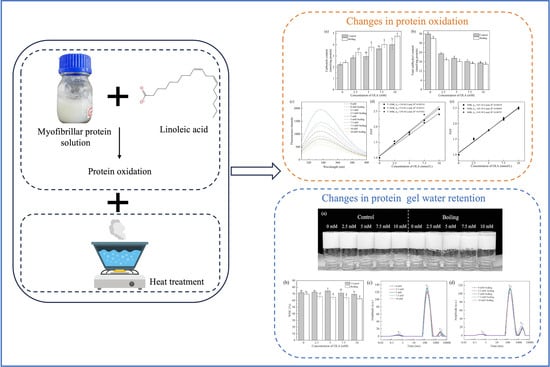
Therefore, grass carp MP was studied in this paper, and the heat treatment was applied following OLA-induced MP oxidation to investigate the impact and possible mechanisms of the heat treatment on the oxidation of grass carp MP. The changes in MP oxidation, thermal aggregation, and gel properties were analyzed by measuring the carbonyl and sulfhydryl contents, surface hydrophobicity, solubility, and WHC. The results may contribute to a deeper understanding of the oxidation mechanisms of MP under the dual influence of heat treatment and OLA oxidation, providing scientific guidance for protein protection and quality control during the heat processing of fish.
2. Materials and Methods
2.1. Samples and Materials
The grass carp were purchased from Yonghui Supermarket in Chongqing, and muscle tissues were collected and portioned into packages of 100 g each at temperatures between 0 and 4 °C. The linoleic acid was chromatographically pure (Macklin, Shanghai, China). 2,4-dinitrophenylhydrazine (DNPH), trichloroacetic acid (TCA), guanidine hydrochloride, and 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB) were purchased from the Aladdin Chemistry Co., Ltd. (Shanghai, China). Tris-(hydroxymethyl)-aminomethane (Tris), tetramethylenediamine (TEMED), ammonium persulfate (APS), and glycine were obtained from the Sigma Chemical Co. (St. Louis, MO, USA). Coomassie brilliant blue R-250 was purchased from the Labgic Technology Co., Ltd. (Beijing, China). Bromophenol blue (BPB) and sodium dodecyl sulfate (SDS) were purchased from Kelong Chemical Reagent Co., Ltd. (Chengdu, China).
2.2. Experimental Methods
2.2.1. MP Extraction
The extraction of MP was conducted by the method of a previous study, with slight modifications [
31]. The grass carp meat (10.00 g) was added to 40 mL of pre-cooled 0.01 M phosphate buffer (containing 0.1 M NaCl, 0.002 M MgCl
2, and 0.001 M EGTA, pH = 7.0) and homogenized at 10,000 rpm for 2 min. The sample was then centrifuged at 4000×
g for 10 min at 4 °C. This process was repeated twice. The resulting precipitate was then resuspended in 40 mL of pre-cooled phosphate buffer (pH = 6.25) containing 0.1 M NaCl, homogenized at 10,000 rpm for 1 min, and centrifuged at 4000×
g for 10 min. This was repeated twice. The final centrifugation was performed by adding 40 mL of pre-cooled phosphate buffer containing 0.1 M NaCl (pH = 6.0) to the last precipitate, homogenizing at 10,000 rpm for 1 min, filtering through four layers of gauze, discarding the precipitate and retaining the supernatant, and centrifuging at 8000 rpm for 10 min. The resulting precipitate was MP. The extracted MP was dispersed into 0.02 M phosphate buffer solution (pH = 6.0, containing 0.6 M NaCl). It was placed in storage at 4 °C and used within 48 h.
2.2.2. MP Oxidation
The MP oxidation method was referenced and modified [
21]. Different concentrations of OLA, 0, 2.5, 5, 7.5, and 10 mM, were added to the MP resuspension solution (10 mg/mL). The samples were incubated at 25 °C in a dark environment for 12 h. After the specified time, the protein was centrifuged at 4 °C for 15 min at 8000 rpm. Subsequently, the MP concentration was adjusted to the desired level and incubated in a boiling water bath for 5 min. The un-boiled MP was recorded as the control, and the 100 °C boiling water bath MP was recorded as the boiling group.
2.2.3. Determination of Protein Oxidation
Carbonyl Content Measurement
The reaction of the proteins with DNPH was utilized to determine the carbonyl content of the proteins [
17]. The MP solution (0.8 mL, 5 mg/mL) was mixed with 2 mol/L hydrochloric acid (1.6 mL, containing 1% DNPH). The reaction was carried out in the dark at room temperature for 1 h. Then, 2 mL of 40% TCA (
w/
v, 40 mg/100 mL) was added to the mixture to precipitate the proteins, which were incubated for 20 min and then centrifuged at 4 °C for 15 min at 8000 rpm. The precipitates were washed with 2 mL of ethanol:ethyl acetate (1:1,
v/
v) until the supernatant was colorless and clear. Finally, the precipitate was dissolved in 2 mL of phosphate buffer (pH = 6.5, containing 6 M guanidine hydrochloride) and incubated at 37 °C until the precipitate was completely dissolved. The absorbance of the solution at 370 nm was determined. The carbonyl content was calculated using the carbonyl molecular absorbance coefficient of 22,000 M
−1cm
−1, and the result was expressed as nmol of DNPH immobilized per mg of protein.
Total Sulfhydryl Content Measurement
The method used to determine the sulfhydryl content was based on a previous study, with minor modifications [
17]. A total of 1 mL of a 5 mg/mL MP solution was mixed with 9 mL of 0.05 M phosphate buffer (pH = 8.0, containing 8 M urea, 0.6 M NaCl, and 0.01 M EGTA). A total of 3 mL of the diluted solution was mixed with 0.4 mL of 0.001 M DTNB. The reaction was carried out at room temperature and protected from light for 1 h. The absorbance was measured at 412 nm.
where “A412” is the absorbance at 412 nm and “C” is the protein concentration (mg/mL).
Fluorescence Spectroscopy
A previously established method was referenced with slight modifications [
31]. The MP concentration was adjusted to 1 mg/mL, and the fluorescence spectrophotometer was set to scan within the range of 300–400 nm, using an excitation wavelength of 295 nm and excitation/emission slit widths of 5 nm.
2.2.4. Determination of Protein Thermal Aggregation Indices
Surface Hydrophobicity Determination
Surface hydrophobicity was determined according to a previous method [
32], with some modifications. A total of 1 mL of a 5 mg/mL MP solution was mixed with 200 µL of a 1 mg/mL BPB solution and 200 µL of a 1 mg/mL BPB, and 1 mL 0.04 M phosphate buffer (pH = 6.8) was added to the control. Then, they were mixed well and reacted at room temperature for 10 min. After centrifuging at 8000×
g for 15 min at 4 °C, 0.1 mL of the supernatant was diluted with 0.9 mL phosphate buffer, and the absorbance was measured at 595 nm. Surface hydrophobicity was expressed as the amount of BPB binding.
Particle Size and Zeta Potential
The MP particle size (d
50) was determined using a Malvern Laser Particle Sizer 2000 [
33]. Adjusting the MP concentration to 10 mg/mL, the relative refractive index and absorption were set as 1.414 and 0.001, respectively.
The concentration of the MP solution was adjusted to 1 mg/mL, and 1 mL of the MP solution was added to the zeta potential dish at room temperature. The zeta potential was tested with a 90° scattering angle, and the equilibration time was 60 s [
31].
Turbidity and Solubility
Turbidity was measured at an absorbance of 370 nm using a 1 mg/mL MP solution [
31].
Solubility was measured as described by a previous study, with slight modifications [
31]. The concentration of the MP solution was adjusted to 1 mg/mL, and the protein concentration was measured at 562 nm. The MP solution was centrifuged at 8000×
g rpm for 15 min at 4 °C, and the supernatant was collected for further analysis.
2.2.5. Determination of Protein Structure
Ultraviolet–Visible Absorption Spectroscopy
The concentration of MP was adjusted to 0.5 mg/mL, and the UV–visible absorption spectra were measured in the range of 230–340 nm.
Scanning Electron Microscope (SEM)
The MP freeze-dried powder sample was glued to the sample stage, and platinum film was plated with an iron sputterer, with the film thickness of about 100 A. The test conditions were as follows: accelerating voltage was 10 kV, and representative surface microstructures were observed and photographed under 5000x magnification.
SDS-PAGE
The cross-linking extent of the proteins was determined by SDS-PAGE [
22]. The concentration of MP was adjusted to 1 mg/mL and mixed with sample buffer in a 3:1 ratio. The mixture was boiled in a water bath for 5 min, cooled to room temperature, and centrifuged at 8000×
g for 5 min. From the supernatant, 200 µL was taken for analysis. A 12% separating gel and a 5% stacking gel were used. Electrophoresis was carried out at 80 V. After electrophoresis, the gel was stained overnight and then decolorized with decolorizing solution until the liquid was colorless. The protein molecular weight was determined using a protein marker.
2.2.6. Amino Acid Content Determination
The amino acid content was determined according to a previous study, with modifications [
34]. An appropriate amount of freeze-dried powder was mixed with 10 mL of 6 M hydrochloric acid (containing 1% phenol), exposed to a nitrogen gas environment for 1 min, and finally hydrolyzed at 110 °C for 22 h. The hydrolyzed sample was diluted to 50 mL. A total of 1 mL of the above solution was blown dry with nitrogen; then, 1 mL of 0.01 M HCl was added to dissolve it, and it was measured through a 0.22 μm membrane. Finally, the filtered solution was tested by an automatic amino acid analyzer.
2.2.7. Determination of MP Gel Water Retention
Preparation of MP Gel
MP gel was prepared based on a previously established method, with some modifications [
35]. The extracted MP was adjusted to 40 mg/mL with a 0.02 M phosphate buffer solution (pH = 6.0, containing 0.6 M NaCl). After oxidation, MP was bathed at 80 °C for 1 h and cooled at 25 °C; then, the protein gel sample was obtained.
Water-Holding Capacity (WHC)
The WHC was calculated according to Equation (4) [
36]. Approximately 3 g of MP gel was centrifuged at 8000×
g, 4 °C for 15 min, and then the resulting weight was recorded.
where W1 represents the weight before centrifugation and W2 represents the weight after centrifugation.
Low-Field Nuclear Magnetic Resonance (LF-NMR) Proton Relaxation
The T
2 value (transverse relaxation time) of gel water was determined by an LF-NMR imaging analyzer [
37]. The magnetic field strength was 0.5T at 32 °C, and the corresponding proton resonance frequency was 22.4 MHz. The other NMR parameters were as follows: SW = 250 kHz, SF = 23 MHz, RFD = 0.080 ms, RG1 = 10.0 db, P1 = 11.52 µs, DRG1 = 4, TD = 1,125,038, PRG = 2, TW = 3000.000 ms, NS = 8, PT = 23.52 µs, TE = 0.250 ms, and NECH = 18,000.
2.3. Statistical Analysis
Statistical analysis was performed using SPSS (Version 27.0) software, and one-way ANOVA was used for significant difference analysis (p < 0.05 indicates a significant difference). Protein secondary structure analysis was performed by PeakFit v4.12 software and plotted using Origin 2024 software (OriginLab Corporation, Northampton, MA, USA).