1. Introduction
1.1. Motivation
The motivation for this research was from the urgent need to enhance neonatal brain structure analysis using MRI. The existing methods face challenges such as low-intensity contrast and the dynamic nature of brain tissue development, leading to suboptimal segmentation and classification results. This research aimed to contribute a novel approach that would address these challenges, providing a more accurate and efficient solution for neonatal brain image analysis. The goal was to advance the field by introducing innovative techniques and methodologies, ultimately improving our understanding of neonatal brain development and contributing to more effective diagnosis and monitoring of neurodevelopmental disorders.
1.2. Related Work
In recent research related to the segmentation and classification of neonatal brain structures, several notable studies have been discussed:
1.3. Research Gaps
Previous studies in neonatal brain tissue segmentation have made valuable contributions, yet certain research gaps persist. One notable gap is the limited exploration of efficient pre-processing techniques to mitigate motion artefacts in neonatal brain MRI. Additionally, the challenge of distinguishing unmyelinated and myelinated white matter in the segmentation process remains underexplored, with existing methodologies relying on conventional approaches. Furthermore, the need for a fully automatic system capable of classifying a comprehensive range of neonatal brain tissues with minimal human involvement has not been adequately addressed.
This work directly addresses the identified gaps in the related literature. We propose innovative pre-processing techniques to effectively correct motion artefacts in neonatal brain MRI, contributing to an improved data quality for segmentation. Our methodology also tackles the nuanced task of distinguishing white matter that is unmyelinated or myelinated, offering a more refined approach compared to conventional methods. Additionally, our research introduces a robust, fully automatic system that maximizes the classification of neonatal brain tissues, filling a critical void in current practices.
Segmenting neonatal brain tissues presents unique challenges due to the rapid growth processes, motion artefacts, and need for comprehensive classification. Existing methodologies often lack the precision and automation required for a thorough analysis. Our research aimed to bridge these critical gaps by developing an innovative segmentation method using the minimum spanning tree (MST) with the Manhattan distance, addressing challenges related to growth and motion artefacts. Additionally, we integrated advanced classification techniques, coupling the Brier score with the shrunken centroid classifier, to enhance precision and reduce manual interventions. Our objectives included comprehensive tissue identification, minimization of human intervention through a fully automatic system, comparative analysis against existing methods, efficient pre-processing for motion artefact correction, and evaluation of the clinical relevance in neonatal healthcare. In setting these objectives, we aimed to advance the state of the art in neonatal brain tissue analysis, contributing to improved healthcare outcomes and a deeper understanding of neurodevelopmental disorders.
1.4. Contributions
This research paper makes several significant contributions to neonatal brain tissue investigation and classification:
- (a)
Enhanced Neonatal Brain Tissue Segmentation: Our work presents an innovative approach for segmenting neonatal brain tissues, overcoming the challenges posed by rapid growth processes and motion artefacts commonly encountered in neonatal brain MRI. We employ cutting-edge techniques, such as minimum spanning tree (MST) segmentation with the Manhattan distance, to increase the robustness and accuracy of segmentation.
- (b)
Advanced Classification Methodology: We presented a novel classification scheme by coupling the Brier score with the shrunken centroid classifier. This hybrid approach enhances the accuracy of tissue classification, leading to more precise and reliable results. It also reduces the dependency on manual interventions and establishes a foundation for fully automatic segmentation and classification.
- (c)
Efficient Tissue Identification: Our methodology addresses the identification of various neonatal brain tissues, including myelinated and unmyelinated white matter, while mitigating the challenges posed by partial volume effects. By enhancing tissue discrimination, we contribute to a more comprehensive understanding of neonatal brain structures.
- (d)
Minimized Human Intervention: We emphasize the importance of reducing human intervention in the segmentation and classification process. Our system aims to be a fully automatic solution, minimizing the need for manual adjustments and interventions, thus streamlining the workflow and enhancing efficiency.
- (e)
Increased Tissue Coverage: In contrast to previous works that segmented a limited number of neonatal brain tissues, our approach endeavors to classify the maximum number of brain tissues with significantly reduced human effort. This expansion in tissue coverage contributes to a more comprehensive analysis of neonatal brain structures.
In summary, our paper presents an integrated framework that not only addresses the challenges of neonatal brain tissue segmentation and classification but also contributes to the goal of creating a robust, automated system for characterizing neonatal brain structures. These contributions collectively to advancing the state of the art in neonatal brain image analysis, with potential suggestions offered for an improved diagnosis and treatment of neurological disorders in neonates.
3. Implementation and Analysis
The proposed methodology can be effectively implemented using the MATLAB 2013b platform. To evaluate its performance and compare it with conventional methods, we conducted comprehensive experiments and analyzed the results.
3.1. Model and Problem Formulation
From this database, the image I is taken, and each of the processes in neonatal brain image segmentation and classification are explained below.
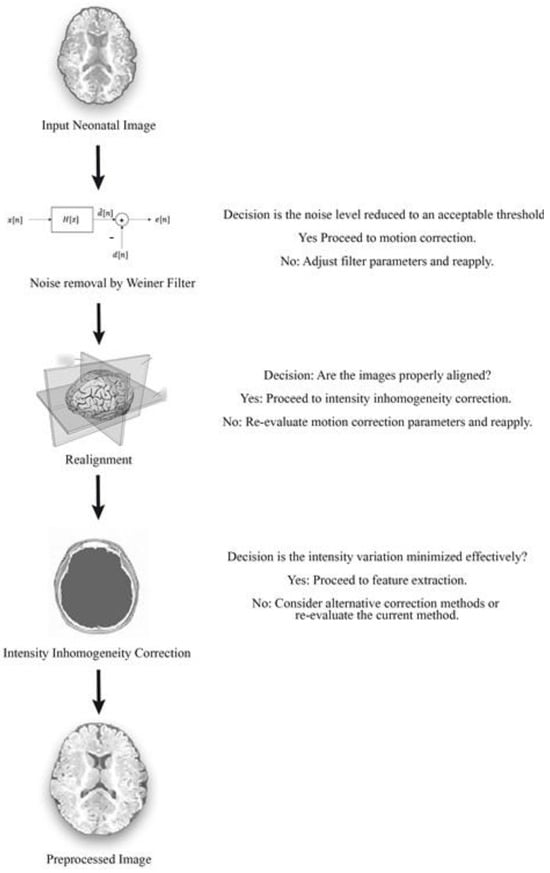
3.2. Pre-Processing (Stage-1)
Pre-processing is the first stage of preliminary processing. It includes cleaning, sampling, normalization, and transformation of the image. The crucial role of pre-processing is to transform the neonatal brain image into a format that will be more easily and effectively processed in subsequent steps. The three pre-processing steps used in our research work were Wiener filtering, realignment, and intensity inhomogeneity correction. The filtering process was used to remove both neonatal brain image noise as well as image blurring. Motion correction was performed in the realignment process and, finally, intensity correction was carried out via the inhomogeneity correction method.
3.2.1. Wiener Filtering
where N = total number of images, = mean of the power spectrum of images N, and = power spectrum of images Ii. By reducing the mean square error of the neonatal brain images, the input images are converged to perfection and hence the noise is reduced. After removing noise from the images, the motion artefacts may be minimized with the help of realignment, and this is explained in the following section.
3.2.2. Realignment
where is a vector function that describes changes in space depending on parameters, = (Pitch, Roll, Yaw), and is the scalar function that describes the intensity transformation depending on parameter = (Pitch, Roll, Yaw).
The intensity of every pixel in the changed image can essentially be resolved from the intensities in the first image. Keeping in the mind the end goal to realign the image through subvoxel precision, spatial changes will include portions of a voxel, resulting in the establishment of a precise correlation among reference and target images. After realignment, the intensity variance in pixels is optimized. Once the correction of motion in the neonatal brain image is completed, the presence of intensity inhomogeneities is removed through the combination of the N3 method and information minimization techniques, as explained in the following section.
3.2.3. Intensity Inhomogeneity Correction
3.3. Non-Parametric, Non-Uniform Intensity Normalization (N3)
This method does not rely on the pulse sequence, and the estimation of dissimilar intensities is shaped by this technique. Bias correction is applied to images of intensities in order to minimize entropy. One advantage of this approach is that it can be used early in an automated data processing procedure, before a tissue model is accessible. For the purpose of calculating intensity variation, an iterative method is utilized to estimate the multiplicative bias field as well as the true tissue intensities’ distribution.
where = the Fourier transform of , = a constant term to limit the magnitude of and U = a distribution function.
The N3 calculation for non-consistency revision makes no presumptions about the intensity of conveyance of fundamental tissue. Instead, U is evaluated non-parametrically from the data by deblurring the deliberate sign intensity V. An assessment of the predisposition field is then derived from the proposed U and y used to adjust the first image. This corrected neonatal brain image is then inputted into the calculation for further redressing. When F or U quits changing with extra cycles, and no more change can be accomplished, the calculation has achieved its objective. After that, information minimization is performed to minimize the impact of intensity inhomogeneity, and this is explained in the following section.
3.3.1. Information Minimization
The information minimization technique is used to minimize or reduce the intensity variation. The elimination of a very higher intensity or very low intensity is based on the average method. In an MR neonatal brain image, intensity varies with respect to location, that is, some regions have a high intensity when compared with neighboring regions, and some regions may have a very low intensity when compared with neighboring regions. This variation in intensity has to be minimized. That minimization is performed by a linear model of image degradation. Initially, we depict the modeling phase, whereby the model’s parametric correction is defined. Second, we describe how to determine the best correction model and calculate the corresponding updated image. An information minimization technique is commonly used in all fields. In this technique, a lower limit and higher limit are set, where the selection of limits is based on the extremity of the intensity. The intensities that are found outside of the limits are minimized to the average value of the limits.
Modelling of Information Minimization Method
where and .
After calculating the inverse degradation model, the intensities must then be corrected, and this is explained in the following section.
3.3.2. Correction of Brain Image
In this equation, “F” represents a noise-reducing filter applied to the product of and . Hence, in the pre-processing section, we reduced the noise level, blurring, motion artefacts, and the intensity inhomogeneity correction, which made the image’s feature extraction more precise. The feature extraction process using DTCWT and the Isomap technique is explained in the following section.
3.4. Feature Extraction (Stage-1)
Neonatal brain MR images offer valuable insights into the structure and development of the infant brain. To extract meaningful information from these images, various features are computed. Textural features capture the spatial arrangement of pixel intensities, providing information about the patterns and structures within the tissue. Intensity features, derived from pixel values, reveal the brightness and color distribution. Shape features describe the geometric properties of segmented regions, such as the area, perimeter, and compactness. Frequency domain features, extracted using the Dual Tree Complex Wavelet Transform (DTCWT), provide insights into the structural characteristics of the images. Finally, statistical features, such as skewness and kurtosis, summarize the distribution of intensities within a region. By combining these features, researchers can gain a deeper understanding of brain development and identify potential abnormalities.
3.5. Dual Tree Complex Wavelet Transform
where are wavelets generated by the two DWTs. Phase and amplitude information is computed using the real and imaginary coefficients. The Dual Tree Complex Wavelet Transform (DTCWT) divides sub-bands into positive and negative orientations using two distinct discrete wavelet transform decompositions. The real and imaginary components are computed through real tree filters (h0 and h1) and fictitious tree filters (g0 and g1), respectively. As shown in Figure 3, DTCWT offers superior directional selectivity and reduced shift variance compared to previous methods. Unlike the critically sampled DWT, which has a 2D redundancy factor for multi-dimensional data, DTCWT provides an improved efficiency with less shift variance and lower redundancy compared to the un-decimated DWT.
| Algorithm 1. for DTCWT |
| 1 Set i = 1 and yield the DTCWT of the input image. 2 Set zero to all wavelet coefficients with the level lesser than a threshold . 3 Proceeds DTCWT-1 and calculate the inaccuracy owing to loss of lesser coefficients. 4 Take DTCWT of the error neonatal brain image and adjust the non-zero wavelet coefficients from step 2 to lessen the error. 5 Increase i, lesson the slightly (to comprise a few more non zero coefficient) and recap steps 2 to 4. 6 When there are enough non zero coefficient to provide the essential rate-distortion trade off, Keep constant and repeat a few extra times until converged. |
Features can vary in relevance: they can be most relevant (containing unique information), relevant (containing some information also found in other features), or irrelevant. Generating a diverse set of features is crucial. Therefore, feature selection is used to select the most relevant features, and here, the Isomap technique is applied.
3.6. Isomap Technique
In the steps involved in the Isomap technique, the neighbor of every point is determined within a fixed radius. The nearest neighbor is taken as k and then we construct a graph by connecting each point, whereas the Euclidean distance is determined by the edge length. By using Dijkstra’s algorithm, Floyd–Warshall algorithms find the shortest distance between two nodes.
The initial step identifies neighboring points on the manifold M, based on distances between pairs of points i and j in the input space X. Subsequently, each point is connected to its K nearest neighbors. These neighborhood relationships are depicted as edges in a weighted graph G over the data points, where the edges have weights based on the distances between neighboring points i and j in the input space X.
where denotes the matrix of Euclidian distances, the τ operator coverts distances to inner products, and . Then, we eliminate the lower dimensional features using the multidimensional scaling method. The obtained feature values are then mapped to the pixel locations present in the image. After the feature extraction process, the image undergoes segmentation.
3.7. Minimum Spanning Tree with Manhattan-Distance-Based Segmentation
Segmentation of newborn brain images refers to the process of dividing or partitioning a digital image into distinct parts or sets of pixels. The main goal of segmentation is that changing the color, intensity, or texture of a desire region so it makes more sense and is simpler to analyze. This segmentation is the process of assigning a label to each pixel in an image so that pixels sharing the same label exhibit specific attributes. Segmentation covers the whole image, where neighborhood regions exhibit substantial variations in similar characteristics. This approach is often employed to detect objects or relevant features within digital images. Common segmentation techniques in image processing include region-based, threshold-based, and edge-based segmentation. But perfect newborn brain image segmentation can be achieved by using the minimum spanning tree with the Manhattan distance. MST segmentation is modified here with the Manhattan distance, which is a dimension-independent distance.
where represents a set of vertices in the graph, and represents a subset of the edges of the graph.
The subset of edges spans all vertices. The distance between two points is classically calculated by means of the Pythagorean Theorem. But one of the effective methods of finding the shortest distance between two points is the Manhattan distance. This is the distance between two points measured along perpendicular axes. In our research, the Manhattan distance was used to find the similar pixels or similar regions in the segmentation process. The map assigns a distance value to each pixel in the image based on its proximity to the nearest obstacle, where pi is the closest pixel causing an obstacle.
The minimum spanning tree technique divides the neonatal brain image up in the form of a grid, and each pixel in the grid is considered the vertex in a graph. A single graph can have multiple spanning trees, each connecting all vertices with the minimum total edge weight. The acquired information from each pixel is stored in the data structure. Kruskal’s MST algorithm is used in the segmentation process, and the edges between the neighborhood pixels are formed then by employing the Euclidean distance, which refers to the distance between two points in Euclidean space. Edge weightage is the measured pixel similarity, which is judged by a heuristic method that compares the weight to a per-segment threshold.
where are points on a plane.
Minimum spanning tree (MST) segmentation using the Manhattan distance is a robust method for partitioning neonatal brain images into meaningful regions. This approach enhances image clarity for diagnostic and research purposes in neurodevelopmental disorders. The Manhattan distance, known for its dimension-independent effectiveness, identifies similar pixels and regions, improving segmentation accuracy. The MST algorithm connects neighboring pixels based on Euclidean distances, with edge weightage determined heuristically. The Manhattan distance, calculated as the sum of horizontal and vertical components, serves as a reliable similarity measure. In summary, the MST with Manhattan-distance-based segmentation is a valuable tool for neonatal brain image analysis, offering enhanced precision and applications in neuroimaging research concerning the gyrus, myelinated white matter, unmyelinated white matter, medial part, lateral part, occipital lobe, cerebrospinal fluid, and temporal gyrus. The overall process involved in the segmentation of newborn brain images can be summarized in the following manner.
3.8. Grid Formation
Divide the pre-processed image U(x) into a grid format where each of the pixels in the image is represented as a vertex in the graph. In the process of grid formation, the information about the pixels is stored in the data structure (DS).
Here, the degree of neighborhood is measured in terms of the feature values obtained in the feature extraction phase. After finding the neighborhood between the nodes, they are sorted, and this is explained in next section.
3.9. Edge Sorting
Edge sorting is a crucial step in image segmentation that involves organizing edges based on their similarity. It facilitates efficient region merging, ensuring accurate and meaningful segmentation.
Procedure:
Weight Calculation: Calculate the weight of each edge using the Manhattan distance.
Sorting Edges: Sort the edges in ascending order based on their weights.
Merging Criteria: Merge regions based on predefined criteria, starting with the lowest-weight edges.
Dynamic Adjustment: Adjust merging criteria based on the image context or analysis requirements.
Edge sorting enhances segmentation quality by ensuring that similar regions are merged first, leading to more coherent and anatomically accurate segmentations. This is particularly important in medical imaging.
In this process, an edge emerges among each pair of nodes; as a result, a set of edges will be formed in the images, and the edges are sorted in ascending order. After that, the merging criterion is checked, and the regions are merged as given in the following section.
3.10. Pixel Merging
Thus, the regions are merged based on Equation (15) if the similarity condition is satisfied; otherwise, merging will not take place. After working through this sequence of steps at all image locations, we will finally achieve a segmented image. Based on the segmented regions, classification of the tissues is performed, and this is detailed in the following section.
3.11. Classification
After segmenting the twelve parts, classification or labeling is needed for easy visualization. The difference between segmentation and classification is that segmentation segregates the neonatal brain image into internal homogenous chunks, but does not specify what the chunks belong to, while classification specifies where each chunk belongs and labels it. Classification of data collected from sensors involves assigning each data point to a corresponding class based on groups with homogeneous characteristics. The goal is to distinguish between different objects within the image. The first step followed in classification of an image is defining the class, and the next is to establish the features that are used in distinguishung the class within the neonatal brain image sampling of training data. Training data or the test data are sampled in order to determine decision rules in the sampling process, or a standard technique is used. The decision rules are confirmed by comparing various classification techniques with the test data. After determining the appropriate decision rule classification is being carried out, all pixels are classified into a single class. In our research, we used a hybrid method of the Brier score coupled with the shrunken centroid classifier. This converges or shrinks similar segmented areas into a class by means of a threshold value. The novelty lies here in combining that classifier with the Brier score, which is the best method for selecting the threshold for the classifier.
3.12. Brier-Score-Coupled Shrunken Centroid Classifier
The group centroid Y for gene g and class k is compared to the overall centroid Y by a method that measures the goodness of a segmented part within a class. This comparison takes into account how a segmented part’s overall centroid differs from a class centroid. It divides this difference by the within-class variance, giving higher weights to segments with stable expressions within the same class.
where = number of possible classes, n = number of instances, = predicted class, and = actual outcome.
Therefore, the lower the barrier score is for a set of predictions, the better the predictions are calibrated, so the value lies between zero and one. For example, if the threshold value is 0.6 and the centroid value of a segmented part is 2.4, by using the shrunken centroid classifier, the value of the segmented parts shrinks or converges to 1.8. After shrinking all centroids, it labels the class by applying the nearest centroid rule. Thus, the neonatal brain image is classified, which is the output. A threshold is set for the normalized differences between the class centroids. If these differences are consistently low for all classes, the corresponding segment is discarded. This process minimizes the number of segments that are incorporated into the final predictive model.
Each class centroid is adjusted toward the overall class centroids by a specific amount, known as the threshold. Each centroid shrinks toward zero by a threshold value. After the centroids are adjusted, the new sample is classified using the standard nearest centroid method. Specifically, if a segment is reduced to zero for all classes, it is excluded from the prediction model. Alternatively, it can be set to zero for all classes except one. After shrinking each segment, the whole newborn brain image is classified. This approach offers two benefits: it enhances classifier accuracy by minimizing the noise impact, and it performs automatic segment selection.
6. Conclusions
In this article, we have presented a comprehensive solution for segmentation and classification of neonatal brain images utilizing Manhattan-distance-based minimum spanning tree (MST) segmentation, coupled with the Brier score and shrunken centroid classifier. Our proposed method excels in accuracy by effectively reducing noise’s impact and successfully predicting classes from a large input dataset. With a strong focus on segmentation and classification, this hybrid approach generates anatomically precise results, thus enhancing subsequent MST-based segmentation.
Through rigorous experimentation, our methodology consistently proved to have a better performance compared to conventional techniques, as set out in the Results section. We achieved remarkable results, including a DSM of 0.8749, CKC of 0.8728, and Jaccard index of 0.9562. These results demonstrate that our methodology surpasses the capabilities of existing conventional segmentation methodologies for neonatal brain images, even in the absence of atlases.
In conclusion, our proposed framework offers a robust solution for neonatal brain image analysis, with the potential to meaningfully impact clinical practice and find research applications in the field of neonatal neuroimaging.
The proposed MST segmentation and Brier-score-based classifier showed significant improvements in neonatal brain MRI analysis, with potential applications extending to adult brain imaging, other medical modalities (CT, PET), and pathological tissue analysis. This approach can also support neurodevelopmental research and may be integrated into AI systems for enhanced diagnostic capabilities across various clinical fields. These findings highlight the potential for a broader impact of the methodology in improving diagnostic precision in healthcare.
Source link
Tushar Hrishikesh Jaware www.mdpi.com