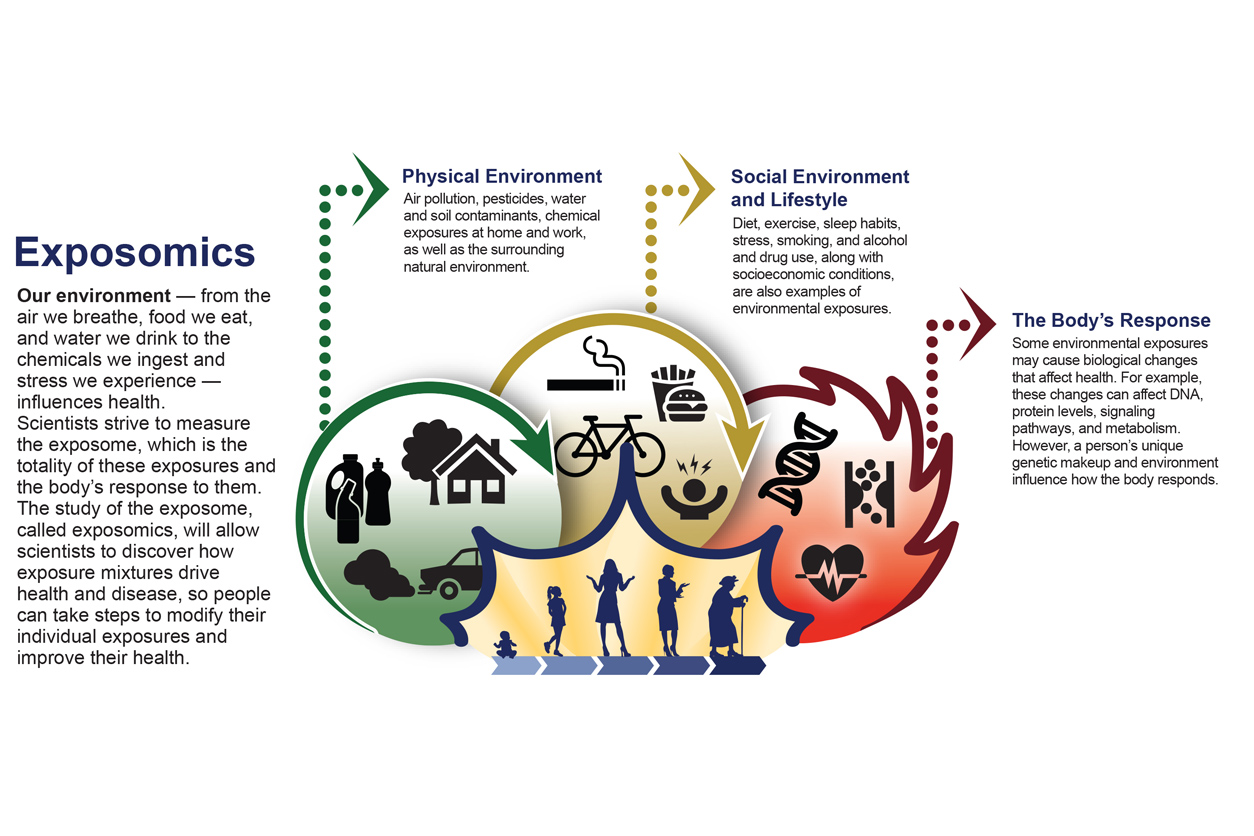
When I launched this column three years ago, one of the first topics I highlighted was the exposome, which refers to the totality of our environmental exposures and their corresponding biological effects. It has become clear to me that the environmental health sciences community should strive to assess how real-world exposures — which almost always involve more than one toxicant — affect human health rather than focusing on just one substance at a time. This is where exposomics can provide a helpful research framework, in my view.
During the past couple of years, NIEHS has made great progress in advancing the relatively new field of exposomics by convening the best and brightest minds in this area, identifying research gaps, and initiating efforts to expand research in the U.S. and around the world.
For example, in 2022, our institute led a successful workshop series titled “Accelerating Precision Environmental Health: Demonstrating the Value of the Exposome.” Precision environmental health is about understanding how the multitude of exposures we experience each day can interact with our unique genetic and biological makeup, and how such interaction may influence health and disease. The goal of precision environmental health is to understand health risks at the individual level and to use that knowledge to prevent disease.
The workshops attracted more than 400 experts and covered topics such as data integration, geospatial technologies that can shed light on the exposome, and standardization of research methods. Out of those meetings, NIEHS sought to develop a community of practice that will help to define best practices and strengthen ongoing efforts through increased scientific partnership and knowledge-sharing. This NIEHS-led community of practice will serve as the U.S. counterpart to ongoing exposomics initiatives in Europe. We are supporting a grant that will create America’s first Center for Exposome Research Coordination, set to launch this summer.
Recently, I sat down with two people at NIEHS who have helped to spearhead these and other exposome research initiatives. David Balshaw, Ph.D., directs the institute’s Division of Extramural Research and Training, and Yuxia Cui, Ph.D., co-directs the NIEHS Human Health Exposure Analysis Resource, which aims to advance exposomics by providing critical scientific expertise and infrastructure. Drs. Balshaw and Cui explained why it is important to study the exposome and shared how NIEHS is leading the way in advancing this research framework. Excerpts of our interview are provided below.
More than one exposure at a time
Rick Woychik: What exactly is the exposome, and why is it an important research framework?
David Balshaw: The exposome concept was originally laid out by Dr. Christopher Wild in 2005, shortly after the Human Genome Project was completed. His argument was that in the same way that the field of genomics had sought to study more than just an individual gene at a time, environmental health sciences needed a similar concept, whereby we assess the totality of exposures that an individual experiences from birth until death. So, the exposome is a kind of continual record of everything you’re exposed to in the environment.
Yuxia Cui: Yes, the exposome is a framework that encourages scientists to embrace the complexity of our environment. It is about getting beyond studying one exposure at a time and instead assessing many real-world exposures, which are numerous and complex.

Framework for scientific discovery
RW: How does one study the totality of environmental exposures? It seems like a concept that may be difficult for scientists to sink their teeth into.
DB: Yes, it is probably not realistic to study every exposure, everywhere, in every person. Over time, the concept has been adapted to more tailored kinds of research questions, and there have been efforts to separate it into more tractable chunks. For example, some scientists focus on the internal exposome, meaning chemicals that can be measured in a biological sample, while others may tackle the external exposome, which might include air we breathe, food we eat, and so forth.
YC: As David mentioned, one task has been to break the exposome into more manageable parts. In some cases, that might involve trying to assess all of the exposures that are correlated with a given disease outcome, to try to determine what specific exposures drive that outcome. Or it may mean conducting hypothesis-free research, where you’re saying that a given health outcome is influenced by something in the environment and we want to look at a broad swath of exposures to identify unknown exposures that underlie that outcome.
DB: Exactly. The exposome is really just a framework for discovery in environmental health.
Capturing real-world exposures
RW: Can you talk about what NIEHS has done — and is doing — to support exposomics?
DB: The approach we’ve taken historically has been to build the scientific capabilities necessary to study the exposome. For example, one of our early efforts involved the Genes, Environment and Health Initiative, which was developed around the same time that Chris Wild was initially laying out the exposomics concept. The initiative was a collaboration between us and the National Human Genome Research Institute. For our part, we focused on supporting the development of wearable sensors to assess air pollution in real-time, and we coupled that with sensor systems looking at diet, physical activity, and psychosocial stress.
YC: Over the years, we have supported the development of cutting-edge exposure sensors through traditional research grants, Small Business Innovation Research grants, and other funding mechanisms. Some of our grantees have developed wristbands that can detect everyday exposures encountered at home, in occupational settings, and so forth. Others have developed wearable sensors to detect different forms of particulate matter. In fact, our institute has collaborated with the National Institute of Diabetes and Digestive and Kidney Diseases on a program to understand the environmental and genetic drivers behind chronic kidney disease of uncertain etiology, and the project is incorporating sensors developed by NIEHS grantees, such as silicone wristbands and air pollution sensors.
Building research infrastructure
RW: I understand that the NIEHS Human Health Exposure Analysis Resource [HHEAR] has also helped to advance exposomics. Can you expand on that resource?
DB: Sure. The idea behind HHEAR is to increase researchers’ access to high-quality exposure assessment capabilities. Among other achievements, we have expanded scientists’ ability to assess environmental samples, whether involving water, soil, or air filters, or wristbands. The program is establishing a database that can integrate different exposures, biological endpoints, and analyses to answer questions that could not be addressed by a single study.
YC: Yes, NIEHS has a long history of supporting exposure science technology, but HHEAR is different because it is about infrastructure — pulling together all of the relevant technologies, data, and expertise so the scientific community can harness them effectively. Similarly, the community of practice that we have sought to develop following our 2022 workshops is all about providing the scientific support necessary for high-quality exposomics studies, and I am excited about the Center for Exposome Research Coordination that will launch later this year.
U.S. innovation and global collaboration
RW: Going forward, what challenges need to be addressed, and what do you hope to achieve?
DB: Technology development will be critical to exposomics. The major technology we have right now for assessing exposures in biological samples is high-resolution mass spectrometry, but there are challenges in its ability to identify compounds. For example, we know that there are 4,000–5,000 high production volume chemicals in commerce. However, the largest studies I am aware of have been able to identify only about half of that amount.
Also, there is a knowledge gap when it comes to being able to use epigenetics to understand how past exposures may be influencing health today. [Epigenetics refers to chemical modifications on DNA or the proteins associated with DNA that affect how genes are turned on and off.] We know that environmental exposures influence epigenetic markers, but do we have the precision to be able to link those markers to a particular exposure at a specific point in time? Going forward, the scientific community will need to be very systematic in terms of mapping what biological signatures are downstream of a historic exposure as opposed to one that is occurring right now.
YC: We have had numerous workshops and meetings with scientists in the U.S. and around the world, and our major goal is to make sure we are working together. For example, the European Union is standing up a coordination center for exposome activities, very similar to the one we are supporting here in the U.S. this year. We’ll also work with professional societies and experts in Asia, South America, and Africa who are trying to bolster their exposomics efforts. Such collaboration is key because this research endeavor cannot be tackled by one country in isolation.
Source link
factor.niehs.nih.gov