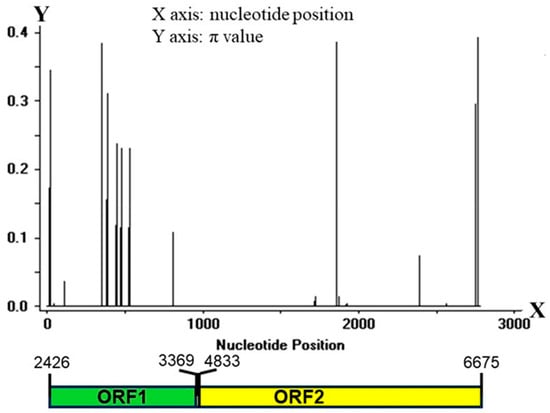
Rice blast is one of the most serious diseases affecting rice crops. Disease resistance breeding is the most economically and ecological friendly way to control this disease. However, most resistance genes have a short life span in the field [9]. Therefore, it is mandatory to select germplasm resources with polygenic resistance for the sustainable control of rice blast. Currently, over 100 blast resistance genes have been identified, and among them the Pi-ta gene stands out as one of the most effective and durable resistance genes. In the United States, the Pi-ta gene has conferred resistance to the major pathogenic forms of M. oryzae [26]. A study has shown that the Pi-ta gene provides 14 years of durable resistance to the contemporary field populations of M. oryzae in rice-growing areas of the southern United States [27]. Some studies have reported the introduction of the Pi-ta gene into cultivated rice varieties from “Tetep” and “Taducan” [10,15,28]. Our results showed that approximately 95% (385 out of 405) of rice landraces collected from different regions in Yunnan Province carried the Pi-ta gene. These samples with the Pi-ta gene could be divided into 28 haplotypes based on the variation analysis of nucleotide sequences in the coding regions and found 12 novel Pi-ta protein products. The haplotype diversity of the Pi-ta gene may be associated with the difference in rice landraces in these different regions. Yunnan in China is known as the origin of multi-cropping agriculture, including rice [21]. This rich history and genetic diversity make it a valuable resource for ongoing research and development in disease resistance for crops such as rice. These results indicate that the haplotypes of the Pi-ta gene are diverse, the variation of nucleotides in its coding region is favorable, and the cultivated rice landraces in Yunnan Province have long held the Pi-ta gene. Thakur and his workers [20] analyzed the variation of Pi-ta alleles in 529 rice landraces in India and in 220 rice accessions, finding that there was a high degree of nucleotide variation in the Pi-ta gene in the intron region, and that 64 Pi-ta haplotypes and 47 Pi-ta protein variants were identified, according to the nucleotide polymorphism of the coding region and amino acid sequences of its locus, respectively. Wang et al. [13] showed that the Pi-ta alleles in rice accessions from 6 Oryza spp. (Oryza sativa, Oryza glaberrima, Oryza rufipogon, Oryza nivara, Oryza Glaberrima, Oryza Rufipogon, and Oryza Nivara) could be divided into 16 different haplotypes. The diversity of the Pi-ta gene haplotypes observed in this study aligns closely with the findings from two previously mentioned studies. The plant R gene polymorphism is an important part of plant innate immune resistance to pathogens, and most R genes are highly polymorphic and diverse [29]. Similarly, avirulence (AVR) genes corresponding to R genes are also in frequent variation. Studies have shown that the AVR-Pita1 gene, an AVR gene from M. oryzae and the interaction with Pi-ta triggering a downstream immune response, is also in a condition of frequent variation [30]. Thus, the diversity of the Pi-ta gene haplotype in different growing regions of rice in Yunnan may also be related to the continuous variation of AVR-Pita1, as the host resistance gene is inclined to continuous variation when the AVR gene form in the pathogen is in a situation of frequent mutation [31,32]. Furthermore, previous studies have shown that the frequency of avirulent gene AVR-Pita1 in 366 M. oryzae rice isolates from different rice regions in Yunnan fields is 46.7–72.4%, and among them, the sequenced 60 isolates code for 18 AVR-Pita1 haplotypes, of which 6 haplotypes are virulent to the Pi-ta R alleles, and the mutations of AVR-Pita1 are responsible for defeating race-specific resistance in nature [33]. Thus, the diversity of the Pi-ta gene haplotype in different growing regions of rice in Yunnan may also be related to the continuous variation of AVR-Pita1, as the host resistance gene is inclined to continuous variation when the AVR gene form in the pathogen is in a situation of frequent mutation [28,30,31]. This represents the coevolution of the AVR gene and the R gene with the host and pathogen interaction. In addition, the continuous variation of AVR-Pita1 may contribute to its unique location. Specifically, AVR-Pita1 is located on chromosome 3 of the M. oryzae genome and is in close proximity to a telomere [34]. The coevolution of the rice gene Pi-ta and M. oryzae gene AVR-Pita1 is still the focus of researchers [15,16,35], probably as the pair of genes strongly supports a hypothesis for gene-to-gene. For instance, Jia et al. [36] showed that rice expressing the Pi-ta gene is resistant to isolates of M. oryzae, expressing AVR-Pita1 in a gene-to-gene manner. Resistant reactions to blast were triggered by the direct interaction of Pi-ta with AVR-Pita1 products, and serine instead of alanine at position 918th in the Pi-ta protein resulted in the functional loss of resistance in plants or the replacement mutation of a single amino acid at position 178th in the AVR-Pita176 protein also disrupted to their physical interaction in vitro. In addition, an immunoreaction mediated by Pi-ta was required to be assisted by the resistance gene Ptr(t) [18,19]. These results suggest that the resistant reaction mediated by the Pi-ta gene is complex in rice.
Genetic evolution might be viewed as a form of biological adaptation to environmental conditions. Lee et al.’s [35] analyses of the genetic evolution of the Pi-ta gene in invasive weedy rice in the United States showed that Pi-ta in weed rice in the United States can be divided into 5 clusters, containing 8 different Pi-ta haplotypes. Only 1 subcluster (a haplotype) in these clusters, however, contained Pi-ta haplotype (Ala-918) resistance to rice blast. In the current study, 35 Pi-ta alleles can be divided into 2 different clusters. Cluster I contained 4 Pi-ta haplotypes (wild O. barthii, O. glaberrima, O. sativa f. spontanea, and O. sativa Indica Group), while Cluster II contained wild Oryza rufipogon and Oryza glaberrima. Furthermore, in 28 Pi-ta haplotypes in rice landraces collected from different regions of Yunnan, apart from 5 haplotypes carrying Pi-ta R alleles, the remaining haplotypes were derived from susceptible plants holding the Pi-ta gene. Analysis of the molecular evolution and functional adaptability of the Pi-ta gene in 36 wild Oryza rufipogon showed that haplotype H2 (Ser-918) is the ancestor of haplotype H1 (Ala-918), and most rice accessions containing the Pi-ta gene belong to H2 in the 26 haplotypes; it was rare that the haplotype H1 would emerge in the process of rice cultivation and domestication and nucleotide diversity of the Pi-ta gene in these rice accessions [35]. Our results showed that H01 (same with EU770212.1) (Ser-918) was the ancestor of the other haplotypes. In general, the variation frequency of nucleotides in the CDS1 and CDS2 regions of the Pi-ta gene was low, and the results are similar to those of previous studies. To investigate the diversity of the exon and intron regions of the Pi-ta gene in 51 rice samples from 6 O. spp., the Pi-ta alleles were classified into 2 main branches, consisting of 16 different sequences with a large number of insertions and deletions [13]. Despite this, the DNA sequence showed 16 variations of the Pi-ta allele in 51 rice samples, but only 9 corresponding protein products were predicted, of which only 1 Pi-ta resistance allele was identified [13]. Their results suggest that the Pi-ta gene has a certain proportion of amino acid synonymous substitution under natural selection, and only a few of them have evolved to carry gene sites resistant to rice blast. In this study, we found 12 novel Pi-ta protein products in 385 rice landraces in Yunnan, of which 5 novel protein products (Ala-918) possessed the ability to recognize AVR-Pita1, and a total of 62 rice landraces coded these proteins. Interestingly, all of the Pi-ta resistance alleles in the rice landraces in Yunnan were derived from the susceptible plants carrying Pi-ta. Furthermore, analysis of Tajima’s neutrality test showed that the Pi-ta alleles could suffer from balancing selection, indicating that the Pi-ta locus probably maintained the diversity of the sequences via the pattern. This information indicated that the Pi-ta gene of rice in Yunnan is constantly subjected to variations, and the Pi-ta gene is always under natural selection pressure; although, it had evolved to the resistance allele. The molecular characteristics of the plant R gene reveal the degree of structural variation that affects its ability to detect the corresponding AVR genes from pathogens [37,38], and the continuous evolution of the Pi-ta gene further indicates that AVR–Pita1 is also in a condition of constant variation. In conclusion, our findings indicate that the Pi-ta gene is present in rice landraces across different rice-growing regions in Yunnan. Furthermore, certain rice accessions with the Pi-ta gene have developed resistant alleles. This discovery significantly contributes to the rich genetic resources available for the investigation of pathogenicity and screening rice accessions for resistance to rice blast.
Source link
Hengming Luo www.mdpi.com