Since the origin and evolution of CeLEA5 genes were not well resolved by above phylogenetic analysis, a similar approach was also used to identify homologs from 33 representative plant species, which include the basal angiosperm Amborella trichopoda, four core eudicots, 25 core monocots, and three early diverged monocots that did not experience the τ WGD, i.e., Acorus gramineus, eelgrass (Zostera marina), and duckweed (Spirodela polyrhiza) [40,41]. As shown in Table S2, a single member was not only identified in A. trichopoda, a rare example without any recent whole-genome duplication (WGD), but also in eelgrass, duckweed, apostasia (Dendrobium catenatum), and Dioscorea alata, though they were proven to have undergone at least one recent WGD after monocot radiation [40,41,42]. By contrast, 2–6 members were found in other species (Table S2), implying the monogenic origin of the LEA_5 family followed by lineage-specific expansion.
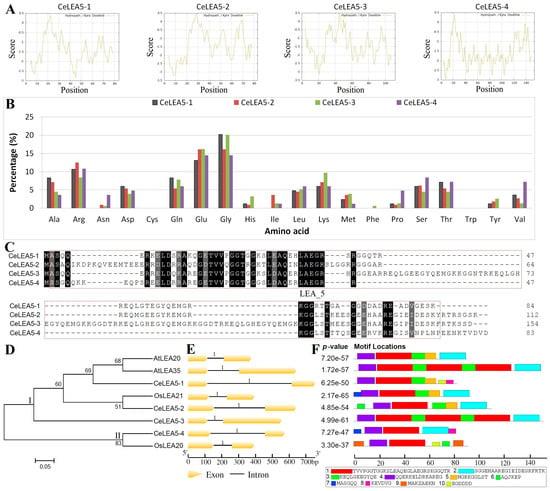
To infer lineage-specific evolution, orthologous genes among different species were clustered using Orthofinder [43]. As shown in Figure 2 and Table S3, a total of five orthogroups were identified. Whereas LEA5a is shared by both monocots and eudicots, LEA5b/LEA5c/LEA5d and LEA5e are specific to monocots and eudicots, respectively. LEA_5 family genes in A. trichopoda, A. gramineus, eelgrass, duckweed, garden asparagus (Asparagus officinalis), and oil palm (Elaeis guineensis) belong to LEA5a, which also includes CeLEA5-1 and OsLEA21 (Table S3), though OsLEA21 was grouped with CeLEA5-2 in the phylogenetic tree as shown in Figure 1D. LEA5b and LEA5d appear to be Poales-specific, whereas LEA5c seems to be confined to Cyperaceae and Juncaceae (Table S3).
To gain insights into the origin of LEA_5 genes, species-specific duplication events were further examined. As shown in Figure 3A, no syntenic relationship was observed between CeLEA5 genes. Instead, CeLEA5-2/-3 and -4 were characterized as dispersed repeats of CeLEA5-1 and -3, respectively, which is similar to that observed in Rhynchospora breviuscula, another Cyperaceae plant. Further interspecific synteny analyses revealed that all CeLEA5 genes have syntelogs in at least one out of 32 species tested in this study, which includes papaya, castor bean, cassava, A. gramineus, and duckweed. Significantly, 1:1 syntenic relationships were observed between tigernut and R. breviuscula, providing direct evidence of early divergence into four groups before Cyperaceae radiation (Figure 3B). Moreover, CeLEA5-1 harbors syntelogs not only in Juncus effuses (a Juncaceae plant within Poales), Sparganium stoloniferum (a Typhaceae plant within Poales), and Joinvillea ascendens (a Joinvilleaceae plant within Poales) (Figure 3C), but also in oil palm, garden asparagus (Figure 3D), A. gramineus, and castor bean (Figure 3E), whereas CeLEA5-2 and -3 have syntelogs in S. stoloniferum/duckweed and J. effuses, respectively (Figure 3C,D). The location of SsLEA5-1, -2, and -3 within syntenic blocks provides direct evidence of LEA5b from LEA5a via WGD, most likely the σ event shared by all Poales plants [44], followed by species-specific expansion via WGD in S. stoloniferum [45]. Notably, though no syntelog was identified for DaLEA5-1 in tigernut, D. alata exhibits 1:1, 1:2, with 1:3 syntenic relationships with duckweed, J. effuses/J. ascendens, and S. stoloniferum, respectively, providing direct evidence of LEA5c from LEA5a via WGD (Figure S1A). Interestingly, in contrast to no syntelog that was identified for all four CeLEA5 genes in A. trichopoda and arabidopsis, A. trichopoda exhibits 1:1 and 1:2 syntenic relationships with arabidopsis/castor bean and A. gramineus (Figure 3E). Moreover, though AtLEA20 and -35 were characterized as dispersed repeats, both of them were shown to be located within syntenic blocks with CpLEA5-1, CpLEA5-2, RcLEA5-1, RcLEA5-2, MeLEA5-1, and MeLEA5-2 (Figure S1B), implying their WGD-derivation followed by species-specific chromosome rearrangement. Interestingly, in contrast to no syntelog that was identified for CeLEA5 genes in all tested Poaceae species, both JaLEA5-1 and -3 were shown to have syntelogs in these species, e.g., Pharus latifolius, rice, and sorghum (Sorghum bicolor). Additionally, tandem duplication was also shown to play a key role in gene expansion of the LEA_5 family in Poales, e.g., J. effuses, J. ascendens, Brachypodium distachyon, barley (Hordeum vulgare), foxtail millet (Setaria italica), and sorghum (Table S2). Notably, despite the occurrence of one additional WGD after the split with sorghum [46], maize has two LEA5a members that were characterized as proximal repeats (Table S2).
Source link
Zhi Zou www.mdpi.com