This research develops a scalable PLB detection and mapping approach using GEE (
Figure 2), enhancing disease surveillance precision. The study created a year-long dataset by integrating Sentinel-1 and Sentinel-2 data, extracting various indices to analyze spatial patterns and complexity by time series dataset. The detection of PLB involves multi-source remote sensing data, which have high-dimensional and complex feature distribution. K-Means clustering can effectively preprocess data and cluster similar samples together, thereby reducing the complexity of the data and identifying the spatial distribution of potatoes, so as to improve the efficiency of subsequent classification. Firstly, K-Means clustering was used to identify the main patterns and structures in the data, so as to provide more representative training samples for Random Forest. As a powerful ensemble learning method, Random Forest is able to process high-dimensional data and automatically select important features, thereby improving the accuracy and robustness of classification. A combination of K-means clustering and RF algorithms was applied for potato identification and PLB detection, with a decision tree model built by field data to boost model accuracy. The model was evaluated using F1 score, the coefficient of determination (R
2) and the root mean square error (RMSE), yielding spatial distribution of PLB and detailed severity maps.
2.4.1. Construction of Features and Indices
In this study, optical remote sensing and radar remote sensing data are used. The indices calculated based on these original bands are as follows.
Selecting appropriate vegetation indices is essential for quantifying plant responses to disease-induced stress, which manifests in various physiological and biochemical changes [
15,
42]. In this study, a variety of vegetation indices, moisture indices, soil and building indices and other relevant indices were comprehensively used to capture and analyze potato growth status and late blight (PLB) occurrence. This includes Normalized Vegetation Index (NDVI) [
43,
44] and Ratio Vegetation index (RVI) [
45], which are used to evaluate the health and growth of vegetation. Water indices, such as the Normalized Humidity Index (NDWI) and Disease Water Stress Index (DWSI) [
44,
46,
47] to detect water conditions and identify disease-related water stress; soil and building indices, such as the Soil Adjusted Vegetation Index (SAVI) [
48] and the Normalized Building Index (NDBI), which are used to consider soil impacts and differentiate urban areas; and other indices, such as Normalized Canopy Chlorophyll Index (NDCI) [
49], were used to estimate chlorophyll content and further analyze the physiological characteristics of vegetation. The use of these indices aims to make full use of multi-band information for remote sensing data, accurately identify and monitor the occurrence and development of PLB and provide a scientific basis for disease management and agricultural decision-making. The indices utilized are enumerated in
Table 2.
Furthermore, radar features, derived from Sentinel-1 SAR imagery, aid in assessing vegetation biomass, moisture content, and surface roughness, which are critical for monitoring crop health and detecting diseases. The study leverages Sentinel-1 SAR imagery’s distinct interaction with Earth’s surface to glean unique insights into surface and vegetation characteristics [
28,
53]. It specifically employs two polarization modes: Vertical Transmit and Vertical Receive (VV) and Vertical Transmit with Horizontal Receive (VH). The backscattering coefficients,
and
, are calculated from the SAR data to evaluate the radar response [
35,
54], defined as the ratio of the backscattered power to the incident power:
Here, the polarization modes are denoted as and , respectively.
In addition, the texture features can quantify the spatial complexity by assessing the statistical relationships between pixel pairs in a given direction [
55]. The Gray Level Co-occurrence Matrix (GLCM) method is an effective technique for texture analysis [
56]. The GLCM was employed to extract texture indices, such as contrast, variance, mean, and difference variance, which enhance the analytical capabilities of images and provide essential supplementary information for land cover classification and vegetation assessment. Utilizing the robust computational power of the GEE platform with a 3 × 3 window size and a 0° orientation, these indices were calculated for obtaining a set of features that characterize the texture of the study area.
2.4.2. A Potato Identification Model Based on Multi-Source Time-Series Remote Sensing Features and Clustering Algorithms
To improve the PLB identification accuracy, the study utilized the K-Means clustering algorithm to identify potato growing areas because of its ability to efficiently process large-scale data by integrating multi-source time-series remote sensing features in an unsupervised clustering task. The K-Means clustering algorithm is a distance-based clustering method [
57,
58,
59,
60]. The algorithm functions through a recursive reduction of the Euclidean distance between each datum and its respective cluster centroid. Its widespread application is attributed to its simplicity, intuitive design, and efficacy. The objective is to segregate data points into clusters in a manner that optimizes for minimal intra-cluster variance, which translates to minimizing the aggregate Euclidean distances from each data point to the centroid of its designated cluster. The objective function can be expressed as:
where is the objective function, is the number of clusters, which is the set of points in the -th cluster, and is a data point within the cluster, which is the center of the -th cluster, representing the Euclidean distance.
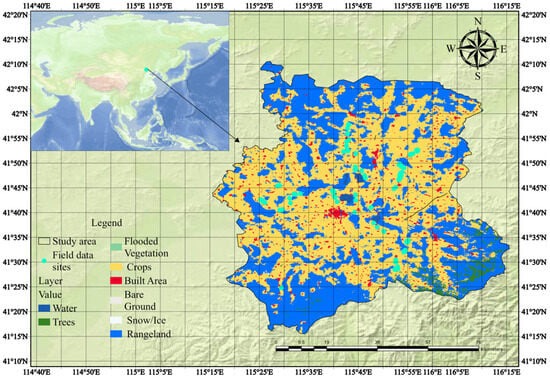
In order to efficiently utilize this clustering method, we created a multi-temporal dataset spanning from January to December 2021 using Sentinel-2 optical and Sentinel-1 radar images. The GEE platform facilitated pre-processing, including noise reduction and smoothing. NDVI and VH band data were extracted to quantify vegetation health and crop structure. These indices were merged into a multi-band image for K-Means clustering, which analyzed 10,000 randomly selected pixels to identify potato-growing areas. The results were validated with 221 field samples labeled as potato or non-potato areas. This integrated approach provides a robust framework for precision agriculture.
In this study, K-means unsupervised classification was first employed to identify potato planting locations, thereby enhancing the accuracy of subsequent machine learning regression. To avoid errors from the initial unsupervised method affecting the accuracy of the subsequent machine learning regression, we implemented several measures to ensure the reliability of our results.
We used the K-means clustering method to identify potato-growing areas by integrating multi-source time-series remote sensing features with field sample data. Morphological operations were then performed on the clustering results to optimize isolated, broken pixels (which are unlikely to represent entire fields). Using the GEE platform, we processed and analyzed field grid images through a series of image processing and morphological analysis methods to identify and optimize crop distribution. Specifically, we defined a 5 × 5 structural element (K) and applied morphological opening operations, including erosion and dilation, to each pixel to remove noise from the image. Gaussian filter is applied to smooth the image after open operation to further reduce the noise. Then, a morphological closure operation is performed to connect adjacent areas by first expanding and then corroding. After that, the number of pixels in each connected region of the image after the closed operation is calculated to extract the features. Finally, the area threshold T is set to keep only the area greater than or equal to T, thereby streamlining and optimizing the classification of farmland areas. This method effectively improves the accuracy and efficiency of farmland distribution identification. In addition, we carried out a detailed error analysis on the clustering results of the K-Means method and selected four real scenes from Google Earth images to illustrate the spatial details of classification, so as to identify possible areas of classification error. Finally, a result-oriented test is carried out. In the final selected model, the correctness of the model is evaluated on the independent verification set, assuming that there are classification errors that may be generated by the K-Means method.
2.4.4. A PLB Identification and Spatial Visualization Model Based on ML and Time-Series Remote Sensing Features
In recent years, machine learning (ML) methods have been extensively applied to address the issue of PLB [
11,
14,
17,
61,
62]. At the same time, unlike complex deep learning models that require a large number of training samples and parameters, GEE has some convenient built-in ML algorithms that allow users to access and process petabytes of remote sensing data. Using these advantages to extract phenological information from time series remote sensing data, favorable insights into the development of different diseases can be obtained from phenological data extracted from each crop growth cycle [
34,
63,
64].
The course of PLB has obvious seasonal and temporal dynamics [
65]. By using time series data, changes in the disease at different stages of growth can be captured [
41,
66,
67], allowing for more accurate prediction and monitoring of PLB severity. Sentinel-1 SAR data and Sentinel-2 optical data each carry unique information. SAR data can obtain surface information under cloudy and rainy conditions, while optical data provides rich spectral information [
68]. By fusing these two types of data, multi-dimensional characteristics of potato growth and disease development can be more fully captured. In particular, PLB changes manifest differently in the early and late stages of the disease, and these changes can be better identified through time series analysis of multi-source data [
43,
69,
70]. Random Forest (RF) [
71] is a powerful ensemble learning method capable of processing high-dimensional data and automatically selecting important features. It performs well in the processing of time series data and can effectively reduce overfitting and improve the generalization ability of the model. Therefore, we propose MSTS–RF (Time Series Random Forest with Multi-Source Data) method. In order to verify the reliability and superiority of the model, we will conduct several controlled experiments, specifically. The first is to verify the superiority of different ML models in PLB problems, and find the most effective model to solve PLB monitoring; the second is to compare the performance of the model without using time series data and the model with time series data; the third is to compare the performance differences between the two single time series data models of radar and optical data and the multi-source time series models, respectively. Finally, the optimal model is selected as the monitoring model for this study. The specific model introduction and modeling process are as follows:
Classification and Regression Trees (CARTs): CARTs employ decision trees for both classification and regression tasks [
72]. In this study, we use the ‘ee.Classifier.smileCart()’ method on the GEE platform to create a CART classifier. In order to optimize the hyperparameters, we search in the range of ‘maxNodes’ (maximum number of leaf nodes) and ‘minLeafPopulation’ (minimum number of samples per leaf node) through grid search and finally determine the number of ‘maxNodes’ to be 10. ‘minLeafPopulation’ is 1 as the best parameter. Training data sets account for 70% and validation data sets account for 30%. The input features include various vegetation indices (such as NDVI, NDWI, NDBI, SAVI, IBI, RVI, DVI, NDCI, DWSI2, DWSI3, DWSI4) and texture features (such as contrast, variance, and standard deviation) calculated based on Sentinel-2 images synthesized one week before and after the date of 2021-08-12, as well as the VV and VH bands from Sentinel-1. The label attribute ‘PLB_percen’ indicates the severity of PLB. During model training, we use root mean square error (RMSE) and coefficient of determination (R
2) to evaluate model performance.
Gradient Tree Boosting (GTB): This ensemble technique progressively incorporates trees to rectify the residuals of the preceding model, directed by the gradient of the loss function [
73]. In this study, we use the GEE platforms ‘ee.Classifier.smileGradientTreeBoost()’ method to create a GTB regression model. The training process is similar to CART. Training and validation are separated, with 70% of the data used for model training and 30% for validation.
Random Forest (RF): An ensemble technique that constructs multiple decision trees and amalgamates their predictive outcomes [
71], introducing randomness to avoid overfitting and enhance accuracy. In this study, we use the GEE platform’s ‘ee.Classifier.smileRandomForest()’ method to create a GTB regression model. The training process is similar to CART. Training and validation are separated, with 70% of the data used for model training and 30% for validation.
Time Series Random Forest Regression with Single Optical time series data (TS–RF): A specialized RF model that uses time-series data from a single source, Sentinel Optics, to analyze and predict trends based on temporal patterns. Using Sentinel-2 satellite data, vegetation and surface indices, such as NDVI, NDWI, NDBI, SAVI, IBI, RVI, DVI, NDCI and DWSI, were calculated, and rich surface features were extracted. In addition, the spatial information is enhanced by GLCM texture analysis. A multi-band time series dataset was established by synthesizing images month by month from May to September 2021. The image features were then trained using an RF model for monitoring PLBS. In the training phase, the data set is randomly divided into a training set accounting for 70% of the data and a validation set accounting for 30% of the data to evaluate the performance of the model. In order to optimize the hyperparameters, we search in the range of ‘numberOfTrees’ (the number of decision trees to create) through grid search, and finally determine ‘numberOfTrees’ 10 as the best parameter. The label attribute ‘PLB_percen’ indicates the severity of PLB. During model training, we use root mean square error (RMSE) and coefficient of determination (R2) to evaluate model performance.
Time Series Random Forest Regression with radar time series data (STS–RF): A specialized RF model that uses time series data from a single source from radar to analyze and predict PLB trends based on time patterns. It is similar to the TS–RF model in construction, but only the VV and VH bands synthesized every month are used for training to explore the disease monitoring capability of the radar.
Time Series Random Forest Regression with Multi-source Data (MSTS–RF): This study presents a Time Series Random Forest Regression model with Multi-source Data (MSTS–RF) to monitor PLB severity using remote sensing data. The dataset consists of 200 bands synthesized from Sentinel-2 and Sentinel-1 satellite data, covering all Sentinel-2 bands, remote sensing indices (e.g., NDVI, NDWI, NDBI, etc., see
Table 2), texture features from GLCM, and radar polarization bands (VV and VH). Monthly image composites were generated for May-September on a month-by-month basis to create a multi-band time series dataset. The training data, labeled with PLB severity classes (0–100%) based on field surveys (221 samples), were partitioned into a 70% training set and a 30% validation set. The Random Forest model was configured with the following hyperparameters: 1000 trees, default variables per split (square root of total variables), minimum leaf population of 1, and a bag fraction of 0.5. The maximum number of nodes per tree was not limited, and the random seed was set to 0 to ensure repeatability. Model performance was evaluated using R² and RMSE. The MSTS–RF outputs continuous values representing PLB severity and spatial distribution, visualized as image layers with legends for reference.
Time Series Gradient Tree Boosting Regression with Multi-source Data (MSTS–GTB): In order to verify the performance of multi-source time series data in different machine learning models, it is proposed to refer to the MSTS–RF model training method, which is consistent except for the different machine learning methods based on it. Using ‘ee. Classifier.smileGradientTreeBoost (10)’ training, we utilized R2 to assess model fit and RMSE to measure the deviation between model predictions and actual data.
Finally, the optimal model is selected comprehensively through the verification performance of these seven models.
2.4.5. Performance Metrics
To evaluate the effectiveness of the proposed models and methods for detecting and monitoring PLB, several performance metrics were employed. These metrics provide quantitative measures of model accuracy, precision, and overall predictive power, allowing for a comprehensive assessment of the model’s performance. The key performance metrics used in this study include , , , , Root Mean Square Error (RMSE), and Coefficient of Determination (R2). Their calculations are as follows.
: refers to the ratio of the number of samples correctly predicted by the model to the total number of samples.
: refers to the proportion of plants that the model predicts to be affected by late blight that are actually affected.
: refers to the proportion of plants that the model correctly detects for late blight. It tells us how successfully the model can predict how many plants will actually be infested.
: The F1 score is a blended average of precision and recall, which combines the accuracy and recall of the model.
Here, (True Positive) is the number of true examples, (True Negative) is the number of true negative examples, (False Positive) is the number of false positive examples, and (False Negative) is the number of false negative examples.
The R-square (coefficient of determination), Root Mean Square Error (RMSE) and Mean Square Error (MSE) were chosen to evaluate the goodness-of-fit of the regression model as follows:
where is the observed value, is the model prediction, and n is the sample size.
To intuitively evaluate the performance of the selected optimal model under different training set sizes and prevent overfitting, we generated learning curves by randomly selecting samples of varying proportions from the original data to form training and validation sets. We tested several ratios, including 90% training and 10% validation, 70% training and 30% validation, 50% training and 50% validation, 30% training and 70% validation, and 10% training and 90% validation. For each ratio, the raw data were divided into training and validation sets, and the selected optimal model was trained on each training set size. We then calculated the performance metrics—root mean square error (RMSE) and coefficient of determination (R2)—for both the training and validation sets. RMSE measures the difference between the model’s predicted values and the actual values, while R2 indicates how well the model fits the data. Using Python 3, we plotted learning curves with the number of training samples on the horizontal axis and the performance metrics (RMSE and R2) on the vertical axis, drawing separate curves for the training and validation sets.
Source link
Zelong Chi www.mdpi.com