1. Introduction
This research introduces an approach to enhance lesion segmentation in oral mucosal disease images through advanced deep learning techniques. By employing sophisticated algorithms, this study uniquely automates the extraction of lesion-specific features, facilitating precise, pixel-level segmentation across diverse types of oral mucosal diseases. Anticipated to markedly boost diagnostic accuracy and streamline treatment processes, this method stands out for its potential to assist clinical practices, showing good clinical application value and prospects.
2. Materials and Methods
2.1. Research Materials
2.1.1. Data Source and Selection
A total of 838 images were collected, including 523 images of OLP, 201 images of OLK, and 114 images of OSF. The image resolution covers three specifications: 8256 × 5504, 6192 × 4128, and 6000 × 4000. The dataset was randomly divided into training, validation, and test sets in a 6:2:2 ratio.
2.1.2. Lesion Site Annotation
All training data were annotated by experienced oral physicians, including those of OLP, OLK, and OSF. Using the LabelMe software (v5.5.0), the disease contours were accurately marked with smooth and continuous curves, and the corresponding diagnosis results were recorded.
2.1.3. Consistency Check and Annotation
Data Annotation and Consistency Validation: Two experienced clinicians independently annotated the acquired training data, delineating diagnoses and disease boundaries for each case, followed by a comparative analysis and consistency assessment. In instances of diagnostic uncertainty, senior consultants were consulted to establish definitive clinical diagnoses, thereby ensuring inter-observer reliability. To assess intra-observer reliability, the same clinicians re-annotated all training data after a two-week interval. Datasets that successfully passed the consistency validation were deemed suitable for inclusion in the machine learning model development. Cases failing to meet consistency criteria underwent additional annotation by a third experienced clinician. Persistently inconsistent datasets were excluded from this study to maintain data integrity.
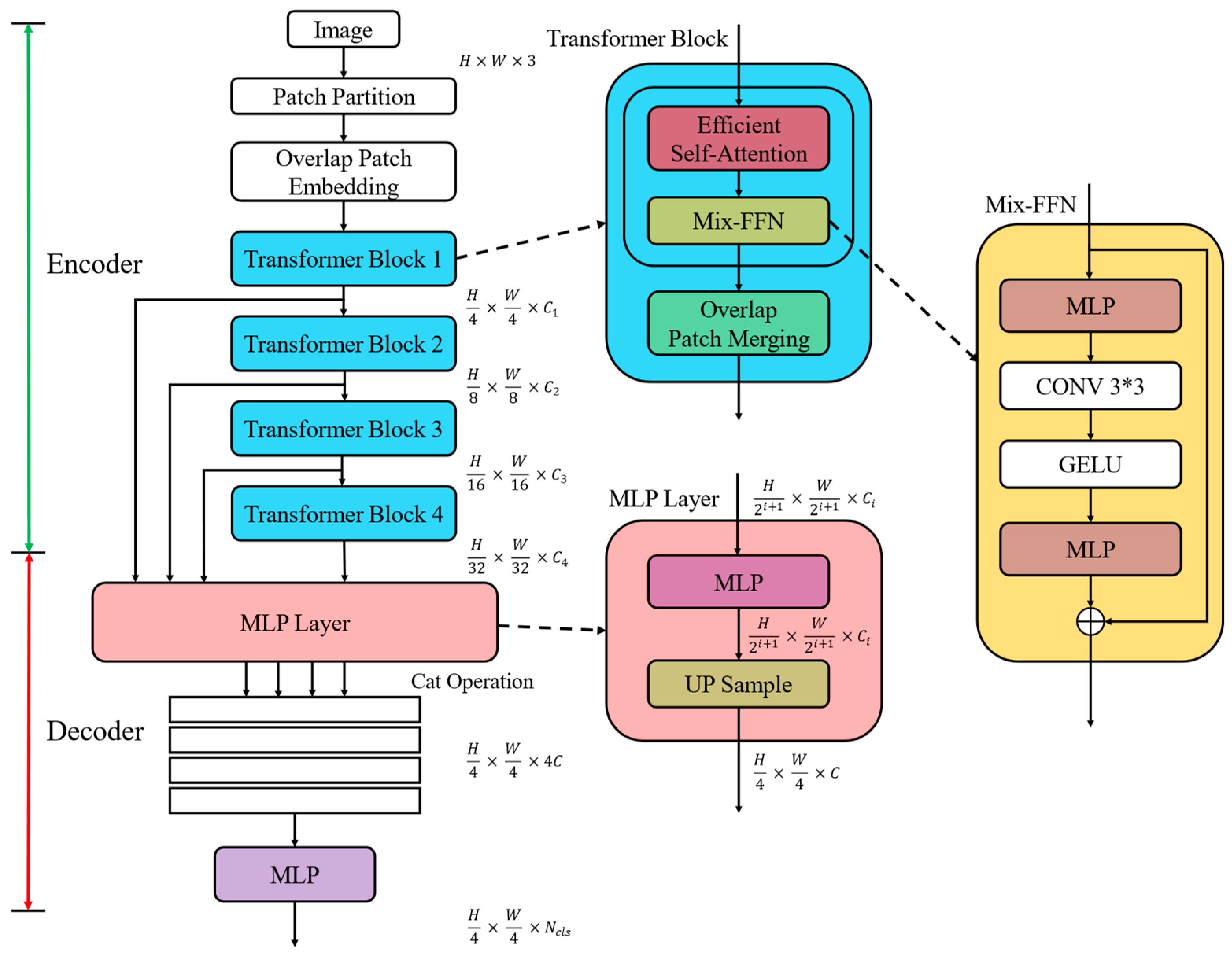
2.2. Construction of SegFormer Semantic Segmentation Model Based on Transformer
2.2.1. Encoder Design
Image patch partitioning—Dividing the input image into multiple equal-sized patches, typically 4 × 4 pixels each;
Sequence processing—Converting image patches into a sequence of vector representations through learnable linear mappings;
Problem transformation—Recasting computer vision tasks as sequence input problems, leveraging Transformer’s global information modeling capabilities;
Position encoding—Introducing position information for image patches to enhance spatial awareness and improve segmentation accuracy.
This design effectively combines the Transformer’s strength in sequence processing with its ability to handle 2D image data, making it particularly suitable for complex semantic segmentation tasks such as oral mucosal lesion detection.
SegFormer enhances the Swin Transformer’s patch merging operation by incorporating an overlapping patch merging technique. This approach allows for pixel overlap between adjacent patches, effectively maintaining pixel continuity and achieving a better balance between global information fusion and local detail preservation.
K′ = Reshape (N/R, C∙R) (K),
K = Linear (C∙R, C) (K′)
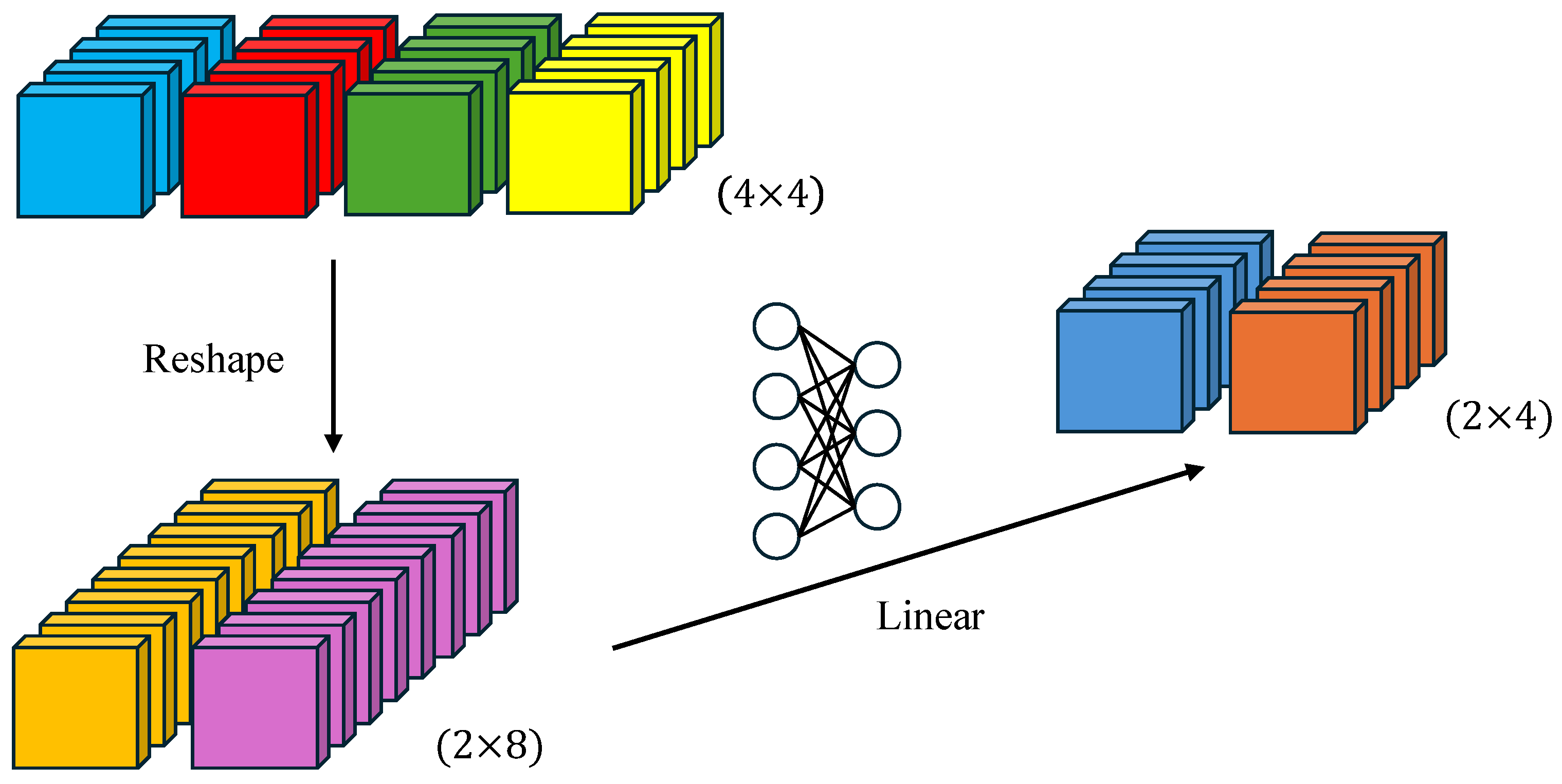
The two-step process first adjusts the input sequence K from N × C to N/R × (C∙R), then projects it back to N/R × C through a learnable linear layer. This reduces the computational complexity of self-attention from O(N2) to O(N2/R), significantly boosting efficiency.
2.2.2. Decoder Design
Fi′ = Linear (Ci, C) (Fi),
Fi″ = Upsample (H/4, W/4) (Fi′),
F = Linear (4C, C) (Concat (Fi″)),
M = Linear (C, Ncls) (F)
This process includes the following:
Feature projection (Equation (3))—The multi-scale feature representations, initially possessing diverse channel dimensions, are linearly transformed to a channel number C. This process facilitates the standardization of feature dimensionality across the different scales.
Feature upsampling (Equation (4))—Feature maps with identical channel numbers but differing resolutions are upsampled to a uniform size of one-quarter of the original image dimensions (H/4 × W/4), ensuring spatial resolution consistency.
Feature fusion (Equation (5))—Feature maps of equivalent resolution and channel count are initially concatenated along the channel dimension. Subsequently, a linear layer projects the channel count back to C, facilitating the effective integration of multi-scale features.
Output generation (Equation (6))—A linear layer projects the channel dimension of the fused feature map to the number of categories Ncls, yielding the final segmentation mask with dimensions (H/4 × W/4 × Ncls).
A decoder based on multilayer perceptrons (MLPs) can simplify model design and reduce computational complexity compared to convolutional neural networks. This MLP-based architecture is able to effectively integrate multi-scale feature information, leading to improved segmentation accuracy. Additionally, the MLP structure is more computationally efficient than complex convolutions, making it easy to adjust and optimize for specific tasks.
The MLP-based decoder design is particularly well suited for the fine-grained segmentation of oral mucosal lesions. By leveraging the multi-scale features extracted by the encoder, this approach can generate high-quality segmentation results. In this way, the SegFormer model is able to achieve efficient semantic segmentation while maintaining strong performance.
Overall, the simplicity and flexibility of the MLP-based decoder, combined with its ability to exploit multi-scale representations, make it a promising architecture for medical image analysis tasks like lesion segmentation.
2.2.3. GELU Activation Function
GELU(x) = xP(X ≤ x) = xΦ(x)
where Φ(x) is the cumulative distribution function of the standard normal distribution. In practice, an approximation is used for computational efficiency.
GELU(x) = 0.5x(1 + tanh[(2/π)1/2 (x + 0.44715x3)])
The GELU function enhances the model’s expressive ability, helps alleviate the gradient vanishing problem, and introduces a slight regularization effect. Its adaptability to inputs of different scales is particularly beneficial for processing multi-scale features in the SegFormer model.
By incorporating the GELU activation function, SegFormer improves feature extraction effectiveness and overall model performance while maintaining model complexity. This is crucial for accurately segmenting lesions in oral mucosal diseases, enabling efficient semantic segmentation with high precision.
2.3. SegFormer Training Configuration
The encoder vector dimension serves as an indicator of the model’s feature extraction capability following each downsampling operation. While the post-downsampling resolution remains consistent across different models, the variation in channel numbers reflects their differing capacities to capture feature complexity. The module depth denotes the number of stacked Transformer modules within the encoder. It is noteworthy that increasing the stacking depth does not alter the feature map dimensions; rather, it reduces resolution and augments vector dimensionality through patch fusion at each module’s output. The number of attention heads influences the diversity of feature extraction, whereas the decoder vector dimension determines the information richness in feature fusion.
These meticulously designed model variants and training strategies are intended to fully leverage SegFormer’s potential in oral mucosal disease segmentation tasks. Through the adjustment of model capacities and the application of efficient training techniques, this study aims to identify the most suitable configuration for this specific task, with the ultimate goal of achieving an optimal balance between accuracy and computational efficiency.
2.4. Data Preprocessing and Augmentation
To enhance the generalization capability and robustness of the SegFormer model in segmenting oral mucosal diseases, a series of meticulously designed data preprocessing and augmentation strategies were implemented. These strategies were devised to simulate real-world image variations, thereby improving the model’s adaptability to diverse image conditions.
The preprocessing phase began with image standardization, normalizing pixel values to the [0, 1] range to mitigate brightness and contrast disparities across images. Subsequently, the following data augmentation techniques were employed:
Random cropping—First, 512 × 512-pixel regions were randomly extracted from original images, encouraging the model to focus on local features and increasing training sample diversity.
Random flipping—Images were horizontally or vertically flipped with 50% probability, promoting the model’s ability to recognize features in various orientations.
Random rotation—Images were rotated within a [−10°, 10°] range, simulating minor variations in capture angles and enhancing the model’s resilience to slight perspective changes.
Brightness and contrast adjustment—Image brightness and contrast were randomly modified within the range of [0.8, 1.2], facilitating the model’s adaptation to varying lighting conditions. Furthermore, random adjustments in hue [−0.05, 0.05], saturation [0.9, 1.1], and color balance [−0.05, 0.05] were introduced to emulate color variations arising from different imaging devices and environments.
These augmentation techniques preserved the original semantic information while substantially expanding the effective training sample size. By introducing these controlled random variations, the model was compelled to learn more robust and generalized feature representations, potentially improving its performance on unseen test data.
This comprehensive data preprocessing and augmentation protocol provided the SegFormer model with a rich, diverse training dataset. Consequently, it mitigated overfitting issues and enhanced the model’s generalization capability and reliability in real-world clinical applications.
2.5. Semantic Segmentation Evaluation Metrics
Dice = 2 × |Prediction ∩ Truth|/(|Prediction| + |Truth|) = 2 × TP/(2 × TP + FP + FN)
In this equation, TP, FP, and FN denote the numbers of true positive, false positive, and false negative pixels, respectively.
IoU = |Prediction ∩ Truth”https://www.mdpi.com/”Prediction ∪ Truth| = TP/(TP + FP + FN)
PA = (TP + TN)/(TP + TN + FP + FN)
These evaluation metrics collectively provide a comprehensive assessment of the segmentation model’s performance, offering insights into its accuracy and effectiveness from various perspectives.
3. Results
The experimental results demonstrate the superior performance of SegFormer models in this task. While the lightweight SegFormer-B0 showed competitive results, SegFormer-B1 and SegFormer-B2 significantly outperformed other models across key performance metrics (Dice, mIoU, and precision). In particular, the SegFormer-B2 model showed marked improvements over established segmentation networks like U-Net and DeepLabV3+, as well as other semantic segmentation models based on Vision Transformer.
The optimal SegFormer-B2 model achieved impressive results with a Dice coefficient of 0.710, mIoU of 0.786, mean pixel accuracy of 0.879, and precision of 0.886. Remarkably, the B2 model achieved these results with a parameter count comparable to or lower than the baseline U-Net while maintaining a computationally efficient profile. Other semantic segmentation models based on Vision Transformer, such as Segmenter and Swin-Unet, generally have a higher number of parameters and computational load. In contrast, SegFormer-B2 utilizes efficient self-attention, which reduces the parameter count while maintaining model performance, thereby offering a clear advantage.
It can be observed that compared to the ground truth annotations in the red areas, SegFormer-B2 provides more precise predictions of lesion edges, offering information about the shape, size, and category of the lesions, which makes it highly referential.
4. Discussion
This study examines the use of the SegFormer deep learning algorithm for semantic segmentation in delineating lesions of oral malignant diseases, providing detailed contours of the lesion areas, which aids doctors in identifying the type, size, and severity of the lesions. This contrasts with other studies in which image classification only provides a general disease category for the entire image without the location information of the lesions—thus offering limited assistance to the diagnostic process. Some studies use object detection algorithms to identify the approximate location of lesions but lack detailed contour information.
The superior performance of the SegFormer model can be attributed to several key factors:
Multi-scale feature fusion—The model’s encoder–decoder architecture enables the simultaneous capture of local details and global semantic information, enhancing segmentation accuracy and comprehensiveness;
Overlapping patch merging—This novel technique maintains local pixel continuity, significantly improving segmentation precision in edge regions and yielding more refined results;
Gaussian Error Linear Unit (GELU) activation function—The incorporation of the GELU provides a regularization effect and enhances the model’s generalization capabilities, allowing for better adaptation to diverse data distributions and task scenarios.
Our experiments comparing U-Net, PSPNet, DeepLabV3+, HRNet, and Vision Transformer-based models and SegFormer models of varying capacities showed that increasing the input resolution and model capacity can promote segmentation performance to some extent. To enhance the coefficient and precision of the model in future applications, it is crucial to collect a larger quantity of high-quality annotated datasets and fine-tune the models. Moreover, further optimizing the model architecture can be achieved by increasing the input image resolution and improving multi-scale feature fusion techniques. Higher input resolution preserves more low-level semantic information, while more sophisticated methods of multi-scale feature fusion can learn more efficiently from features at various resolutions, allowing the model to exhibit superior performance at the same capacity.
Despite these positive outcomes, this study has certain limitations. The dataset, while sourced from a tertiary specialized oral hospital in Zhejiang Province, is relatively limited in scale and geographical diversity. This constraint may impact the model’s ability to comprehensively capture disease characteristics across different populations. Additionally, the lack of external datasets for testing the semantic segmentation model’s generalization limits our ability to fully assess its robustness and transferability. Although the deep learning process included corrections for variations in camera angles and lighting conditions, the absence of validation on external datasets precludes a comprehensive evaluation of the model’s adaptability. Additionally, the model was only trained on three types of oral malignant disease datasets, which may lead to poor or erroneous segmentation for other malignant or benign diseases. Furthermore, the interpretability of deep learning models is subpar, requiring further consideration and research for practical application in clinical diagnostic support.
In clinical applications, automated lesion segmentation in oral mucosal disease images presents a promising approach to enhance oral disease diagnosis and treatment. Compared to subjective manual assessments, this method provides greater standardization and consistency by utilizing objective image features. It can assist clinicians in rapidly and objectively evaluating lesion location, size, and morphological characteristics, potentially detecting subtle abnormalities that might be overlooked by visual inspection alone. This capability could significantly enhance early disease detection and intervention. Moreover, the systematic segmentation of lesion areas across large-scale datasets can contribute to a more comprehensive understanding of oral mucosal disease imaging features, supporting in-depth investigations into disease mechanisms. The quantitative analysis of lesion areas enables the precise monitoring of disease progression, providing valuable insights for treatment planning and adjustment.
5. Conclusions
In conclusion, the SegFormer-based oral mucosal disease image lesion segmentation method developed in this study demonstrates significant performance advantages and broad clinical application potential. As artificial intelligence technologies continue to advance and integrate more deeply with clinical practice, such intelligent diagnostic support tools are expected to play an increasingly crucial role in enhancing the quality and efficiency of oral healthcare, ultimately leading to more precise and effective patient care.
Author Contributions
Conceptualization, J.Z.; methodology, M.L. and J.Z.; software, M.L. and J.Z.; validation, X.C.; formal analysis, M.L.; investigation, Y.C. (Yuqi Cao); resources, F.Z.; data curation, R.Z.; writing—original draft preparation, R.Z. and M.L.; writing—review and editing, Y.C. (Yuqi Cao); visualization, M.L.; supervision, Y.C. (Yaowu Chen); project administration, X.T. and Y.C. (Yuqi Cao); funding acquisition, R.Z., X.T. and Y.C. (Yuqi Cao). All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported part by Science and technology special project, Institute of Wenzhou, Zhejiang University (XMGL-KJZX-202401), Zhejiang Provincial Natural Science Foundation of China (LQ24F030003) and National Natural Science Foundation of China (62303408).
Institutional Review Board Statement
Ethical review and approval were waived for this study due to the reason that this study is a retrospective, non-interventional clinical study that analyzes previously obtained intraoral lesion image data from past diagnoses and treatments. This study does not involve patients’ personal identities or any information that could potentially compromise patient privacy. Furthermore, during the diagnostic and treatment process, subjects agreed that “lesion photographs will be used to observe changes in the condition or treatment efficacy during the treatment and follow-up process, and will be used for summarizing clinical diagnostic and treatment experiences, academic exchanges, and related paper writing and publication, under the premise of protecting patient privacy”.
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data supporting this study’s findings are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jain, N.; Dutt, U.; Radenkov, I.; Jain, S. WHO’s global oral health status report 2022: Actions, discussion and implementation. Oral Dis. 2024, 30, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Liu, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2022, 71, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–27 October 2021. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022. [Google Scholar]
- Keser, G.; Bayrakdar, İ.Ş.; Pekiner, F.N.; Çelik, Ö.; Orhan, K. A deep learning algorithm for classification of oral lichen planus lesions from photographic images: A retrospective study. J. Stomatol. Oral Maxillofac. Surg. 2023, 124, 101264. [Google Scholar] [CrossRef]
- Lin, H.; Chen, H.; Weng, L.; Shao, J.; Lin, J. Automatic detection of oral cancer in smartphone-based images using deep learning for early diagnosis. J. Biomed. Opt. 2021, 26, 086007. [Google Scholar] [CrossRef]
- Welikala, R.A.; Remagnino, P.; Lim, J.H.; Chan, C.E.; Rajendran, S.; Kallarakkal, T.G.; Zain, R.B.; Jayasinghe, R.D.; Rimal, J.; Kerr, A.R.; et al. Automated detection and classification of oral lesions using deep learning for early detection of oral cancer. IEEE Access 2020, 8, 132677–132693. [Google Scholar] [CrossRef]
- Warin, K.; Limprasert, W.; Suebnukarn, S.; Jinaporntham, S.; Jantana, P. Automatic classification and detection of oral cancer in photographic images using deep learning algorithms. J. Oral Pathol. Med. 2021, 50, 911–918. [Google Scholar] [CrossRef]
- Achararit, P.; Manaspon, C.; Jongwannasiri, C.; Phattarataratip, E.; Osathanon, T.; Sappayatosok, K. Artificial intelligence-based diagnosis of oral lichen planus using deep convolutional neural networks. Eur. J. Dent. 2023, 17, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- SM, P.S.; Shariff, M.; Subramanyam, D.P.; Varun, M.H.; Shruthi, K.; Poornima, A.S. Real Time Oral Cavity Detection Leading to Oral Cancer using CNN. In Proceedings of the 2023 International Conference on Network, Multimedia and Information Technology, Bengaluru, India, 1–2 September 2023. [Google Scholar]
- Tanriver, G.; Soluk Tekkesin, M.; Ergen, O. Automated Detection and Classification of Oral Lesions Using Deep Learning to Detect Oral Potentially Malignant Disorders. Cancers 2021, 13, 2766. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Sunny, S.; Uthoff, R.D.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Anbarani, A.; Wilder-Smith, P.; Kuriakose, M.A.; et al. Automatic classification of dual-modalilty, smartphone-based oral dysplasia and malignancy images using deep learning. Biomed. Opt. Express 2018, 9, 5318–5329. [Google Scholar] [CrossRef]
- Yang, S.; Li, S.; Liu, J.; Sun, X.; Cen, Y.; Ren, R.; Ying, S.; Chen, Y.; Zhao, Z.; Liao, W. Histopathology-Based Diagnosis of Oral Squamous Cell Carcinoma Using Deep Learning. J. Dent. Res. 2022, 101, 1321–1327. [Google Scholar] [CrossRef]
- P, S.K.; Lavanya, J.; Kavya, G.; Prasamya, N.; Swapna. Oral Cancer Diagnosis using Deep Learning for Early Detection. In Proceedings of the 2022 International Conference on Electronics and Renewable Systems, Tuticorin, India, 16–18 March 2022. [Google Scholar]
- Deif, M.A.; Attar, H.; Amer, A.; Elhaty, I.A.; Khosravi, M.R.; Solyman, A.A.A. Diagnosis of Oral Squamous Cell Carcinoma Using Deep Neural Networks and Binary Particle Swarm Optimization on Histopathological Images: An AIoMT Approach. Comput. Intell. Neurosci. 2022, 1, 6364102. [Google Scholar] [CrossRef]
- Das, N.; Hussain, E.; Mahanta, L.B. Automated classification of cells into multiple classes in epithelial tissue of oral squamous cell carcinoma using transfer learning and convolutional neural network. Neural Netw. 2020, 128, 47–60. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.; Kim, H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 6994. [Google Scholar] [CrossRef]
- Jeyaraj, P.R.; Samuel Nadar, E.R. Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J. Cancer Res. Clin. Oncol. 2019, 145, 829–837. [Google Scholar] [CrossRef]
- Ye, Y.J.; Han, Y.; Liu, Y.; Guo, Z.L.; Huang, M.W. Utilizing deep learning for automated detection of oral lesions: A multicenter study. Oral Oncol. 2024, 155, 106873. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Sánchez, A.; Bagan, J.; Vila-Francés, J.; Magdalena-Benedito, R.; Bagan-Debon, L. Prediction of the risk of cancer and the grade of dysplasia in leukoplakia lesions using deep learning. Oral Oncol. 2022, 132, 105967. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Chen, Y.; Li, Z.; Jing, Q.; Hu, C.; Liu, H.; Bao, J.; Hong, Y.; Shi, T.; Li, K.; et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. eClinicalMedicine 2020, 27, 100558. [Google Scholar] [CrossRef]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Xu, N.; Li, B.; Liu, Z.; Gao, R.; Wu, S.; Dong, Z.; Li, H.; Yu, F.; Zhang, F. Role of Mammary Serine Protease Inhibitor on the Inflammatory Response in Oral Lichen Planus. Oral Dis. 2019, 25, 1091–1099. [Google Scholar] [CrossRef]
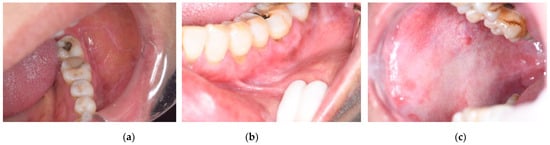
Samples of the dataset. (a) OLP; (b) OLK; (c) OSF.
Figure 1.
Samples of the dataset. (a) OLP; (b) OLK; (c) OSF.
The overall structure of the SegFormer semantic segmentation model.
Figure 2.
The overall structure of the SegFormer semantic segmentation model.
A schematic of the efficient self-attention mechanism.
Figure 3.
A schematic of the efficient self-attention mechanism.
SegFormer semantic segmentation visualization of oral mucosal lesions.
Figure 4.
SegFormer semantic segmentation visualization of oral mucosal lesions.
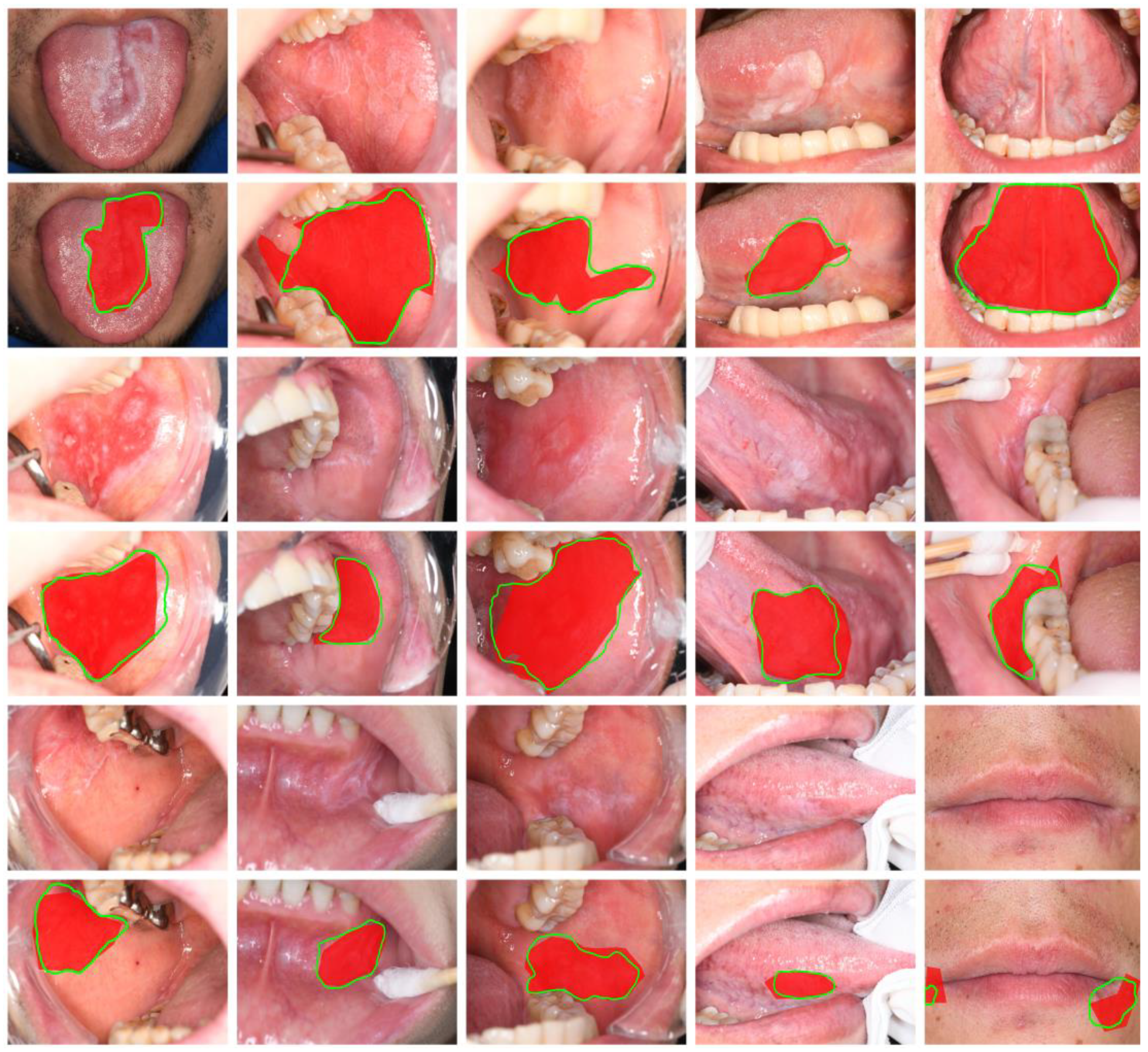
Table 1.
SegFormer model configurations.
Table 1.
SegFormer model configurations.
| Model | Input Resolution | Encoder Dimensions | Encoder Depth | Attention Heads | Decoder Dimension |
|---|---|---|---|---|---|
| B0 | [512, 512] | [32, 64, 160, 256] | [2, 2, 2, 2] | [1, 2, 5, 8] | 256 |
| B1 | [512, 512] | [64, 128, 320, 512] | [2, 2, 2, 2] | [1, 2, 5, 8] | 256 |
| B2 | [512, 512] | [64, 128, 320, 512] | [3, 4, 6, 3] | [1, 2, 5, 8] | 768 |
Table 2.
Semantic segmentation task results.
Table 2.
Semantic segmentation task results.
| Model | Backbone | Params | FLOPs | Dice | mIoU | mPA | Precision |
|---|---|---|---|---|---|---|---|
| U-Net | VGG | 24.82 M | 451.71 G | 0.636 | 0.683 | 0.832 | 0.784 |
| U-Net | ResNet50 | 43.93 M | 184.13 G | 0.665 | 0.728 | 0.867 | 0.813 |
| PSPNet | MobileNetV2 | 2.38 M | 6.03 G | 0.627 | 0.692 | 0.852 | 0.765 |
| PSPNet | ResNet50 | 46.71 M | 118.42 G | 0.670 | 0.725 | 0.862 | 0.819 |
| DeepLabV3+ | MobileNetV2 | 5.82 M | 52.88 G | 0.667 | 0.716 | 0.830 | 0.817 |
| DeepLabV3+ | Xception | 166.85 M | 54.71 G | 0.680 | 0.682 | 0.892 | 0.749 |
| HRNet | W18 | 9.64 M | 32.81 G | 0.669 | 0.737 | 0.853 | 0.824 |
| HRNet | W32 | 29.54 M | 79.93 G | 0.698 | 0.745 | 0.844 | 0.841 |
| UPerNet | Swin-B | 120.20 M | 82.10 G | 0.687 | 0.756 | 0.853 | 0.806 |
| Segmenter | ViT-L | 333.08 M | 665.44 G | 0.704 | 0.752 | 0.865 | 0.758 |
| MaskFormer | Swin-T | 41.76 M | 98.51 G | 0.705 | 0.735 | 0.815 | 0.833 |
| Swin-Unet | Swin-L | 335.80 M | 200.02 G | 0.699 | 0.670 | 0.736 | 0.879 |
| SegFormer | MiT-B0 | 3.72 M | 13.55 G | 0.659 | 0.758 | 0.864 | 0.844 |
| SegFormer | MiT-B1 | 13.68 M | 26.50 G | 0.698 | 0.774 | 0.859 | 0.874 |
| SegFormer | MiT-B2 | 27.35 M | 113.45 G | 0.710 | 0.786 | 0.876 | 0.886 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Source link
Rui Zhang www.mdpi.com