1. Introduction
South Africa is part of the “Elimination 8”, a grouping of eight countries in southern Africa aiming to eliminate malaria [
1]. The elimination of malaria in South Africa is hindered by various factors, including the complex composition of malaria vectors. All the species involved in disease transmission in South Africa are known to bite and rest outdoors. As a result, conventional chemical interventions, such as indoor residual spraying and long-lasting insecticide-treated nets are less effective since they primarily target mosquitoes that bite and rest indoors [
2]. Consequently, the presence of outdoor-biting and outdoor-resting mosquito populations contributes to ongoing low-level transmission of the disease outdoors, referred to as residual malaria [
3,
4]. This is a particular challenge for South Africa’s elimination agenda.
Although current chemical interventions remain effective in South Africa, they are not efficient against outdoor biting vectors. Additionally, insecticide resistance in local mosquito populations presents further challenges to these interventions [
5]. Therefore, alternative and supplementary vector control tools are needed to achieve the goal of malaria elimination. Ideally, these should be a non-chemical intervention. One potential option is transmission blocking, possibly through genetic modification. A promising approach is paratransgenesis, a form of biocontrol where microbial symbionts are used to induce refractoriness in vectors [
6,
7].
Paratransgenesis is based on the manipulation of symbionts, usually bacteria, due to their large influence on the life history of the mosquito. The gut microbiota in particular play a crucial role during the mosquito developmental cycle. The gut microbiota are essential for larval development [
8], with the correct bacterial composition needed for the larvae to pupate [
9]. The gut microbiota are also crucial during digestion, particularly blood digestion [
10]. The bacterial composition is also implicated in insecticide resistance [
11]. The most important factor for paratransgenesis, however, is that gut microbiota are a critical immunomodulator and influence the escape of
Plasmodium parasites from the gut [
12]. The midgut is a major barrier for parasite escape, and if contained in the gut, the chances of transmission blocking are significantly increased [
13]. Gut microbiota modulate the formation of the peritrophic matrix, a chitinous membrane that surrounds the ingested blood meal [
14]. This protects the mosquito from a range of ingested pathogens, including the
Plasmodium parasite [
15]. Microbial components also modulate the immune system to eliminate the parasite [
13]. There are a range of bacterial species that have been identified as potential paratransgenesis candidates. These symbionts are suggested for the control of a range of vector-borne diseases [
16,
17].
Many paratransgenesis candidates are based on their natural capacity to inhibit
Plasmodium parasites. Other species are candidates because of their association with refractoriness and would require genetic modification to improve their anti-
Plasmodium function [
7]. However, not all paratransgenesis candidates are genetically modified. As an example, the
Enterobacter Esp_Z strain identified from Zambian
An. arabiensis is a naturally occurring symbiont capable of inhibiting parasite development [
18]. The microsporidian Microsporidia MB is capable of inhibiting parasite development and was also discovered in
An. arabiensis [
19]. The most used paratransgenesis candidate is the intracellular bacterium
Wolbachia. The most effective use of the symbiont has been where local strains have been used [
20]. As such, successful paratransgenesis efforts require an intensive understanding of the local symbionts where the intervention is to take place.
As mining local populations for potential symbionts is important for this intervention, it is important to understand the importance of bacterial dynamics. It has been recognised that bacterial dynamics differ by season [
21]. Furthermore, it has been demonstrated that the larval environment plays a critical role in adult bacterial composition [
22]. As such, when looking for local symbionts, it is important to understand the effect of the environment on bacterial composition.
Anopheles arabiensis is a member of the
An. gambiae complex implicated in malaria transmission in South Africa [
23]. This species is generally difficult to control [
24] but notably, it has been reported to be adapting to breeding in polluted areas [
25]. This is significant as the
An. gambiae complex typically breeds in clean, clear, sunlit bodies of water [
26]. This has implications for the expansion of the range of this species, but critically, this adaptation has a range of effects on mosquito life history. This includes the selection of insecticide resistance [
27]. As such, adaptation to pollution is important to consider when examining the larval environment’s effect on bacterial composition.
In this study, the effect of larval exposure to heavy metals on bacterial composition and diversity was examined on two laboratory strains of
An. arabiensis. The first is the insecticide unselected primarily insecticide susceptible strain named SENN, and the insecticide selected strain named SENN-DDT. The use of both selected and unselected strains is because it has been previously demonstrated that insecticide resistance affects the gut bacterial composition [
28].
The two laboratory strains in this study have previously been used to examine the effect of larval exposure to heavy metals.
Anopheles arabiensis, like
An. gambiae, has been adapting to breeding in polluted sites, especially metal-polluted water [
29]. The first example of
An. gambiae breeding in polluted water highlighted that the levels of lead, copper, and cadmium in particular were high in the breeding site [
30]. As such, these metals have been the focus of metal adaption studies in the complex [
31,
32,
33]. Copper and cadmium have been demonstrated to have effects on the life history of the SENN and SENN-DDT strains [
34]. This effect lasts to the second generation, even if the adults breed in clean water [
35]. There have also been demonstrations of the effect of these metals on the epigenetic architecture of these strains [
36]. This study therefore expands the study of the effect of pollutants on these two
An. arabiensis strains by examining the effect of these stressors on the gut microbiota. This will be contextualised by a comparison to wild mosquitoes.
2. Materials and Methods
This study compared the dynamics of the effect of larval metal exposure of two laboratory strains of An. arabiensis with that of F1 An. arabiensis from KwaZulu-Natal, South Africa as a wild-type comparator. This will offer insights into how insecticide-resistant phenotypes influence this dynamic. Additionally, it will demonstrate the value of using laboratory strains to understand the environmental impact on bacterial dynamics.
All mosquitoes used in this study were housed in the Botha de Meillon (BDMI) insectary in Sandringham, Johannesburg. Mosquito husbandry procedures were performed as per [
37]. In brief, mosquitoes were reared at 25 °C (±2 °C) at a humidity of 80% (±5%). Larvae were reared on a diet of Beano™ dog biscuits and brewer’s yeast mixed at a ratio of 3:1, respectively. Adults were maintained with a 12:12 h light cycle with a 30-min dawn/dusk cycle. Adults were maintained with ad libitum access to 10% sucrose.
Two laboratory strains of
An. arabiensis were used. SENN is an unselected susceptible strain colonised in 1980 and originates from Gezira, Sudan. This strain had incipient DDT and malathion resistance, which has subsequently declined to near susceptibility [
38]. From this strain, a resistant strain was selected. The SENN-DDT strain is an insecticide-resistant strain selected by continuous exposure to 4% DDT. The selection started in 1995 and continues until the present. SENN-DDT displays resistance to DDT, deltamethrin, λ-cyhalothrin, and malathion [
39]. The resistance in this strain is mediated by elevated enzyme activity of cytochrome P450s, general esterases, and glutathione S-transferases [
40].
The F
1 adults were obtained from adults collected from Mamfene, KwaZulu-Natal, South Africa (S27°20′17.95″; E32°12′53″). Collections took place between November 2019 and April 2020 [
5]. Adults were collected from clay pots and modified bucket traps [
5]. Once transported from the field site to the BDMI, they were kept in quarantine and not allowed any additional blood meals. Females were individually tubed to allow for the generation of isofamily lines. Upon laying a batch of eggs, the females that laid eggs were placed on silica and identified morphologically according to [
41]. Members of the
An. gambiae complex were identified by conventional polymerase chain reaction (PCR) according to the method of [
42]. If identified as
An. arabiensis, samples were pooled and reared according to the standard insectary procedures described previously. Adults used for F
1 screening represent offspring from 50 families. Metal exposures were performed as per [
34]. Samples were exposed to the maximum acceptable toxicity concentration (MATC), as it represented the lowest possible concentration that would qualify as pollution. In brief, for the laboratory strains, the 100 first instar larvae less than 24 h old were placed in 1000 mL water. To this a metal solution was added to a final concentration of either 0.36 μg/L for cadmium chloride or 1.86 μg/L for copper nitrate [
32]. Larvae reared in untreated water served as a control. For the F1 larvae, 50 larvae per 500 mL were pooled at the same concentrations as for the laboratory strains. These experiments were replicated three times. All larvae were fed a set amount of food (1 mg food per larvae, twice daily). To collect the specimens for sequencing, the adults were collected and kept in cages corresponding to their emergence date. Adults were cold killed at the age of three days. During the period prior to their killing, females were not allowed a blood meal. These mosquitoes were used for subsequent DNA extractions.
2.1. Preparation of Samples for Sequencing
The external surface of the samples was sterilised using 70% ethanol. The midguts were resected in a sterile manner at 40× magnification using an Olympus SZ40 (Olympus LS, Tokyo, Japan). Three guts were used per replicate, with five replicates prepared for each treatment. Total genomic DNA was extracted using the Qiagen blood and tissue kit (Qiagen: 69506, Hilden, North Rhine-Westphalia, Germany). The optional RNAse-A treatment was performed using the kit enzyme (Catalogue number: 19101). An extraction control was included in the reaction (Zymo Research: D6300, Irvine, CA, USA).
The V3–V4 hypervariable region of the bacterial 16S rRNA gene was amplified using conventional PCR [
43]. The amplification was performed according to the Illumina 16S Metagenomic Sequencing Library Preparation protocol (Illumina TM, San Diego, CA, USA). Samples were amplified using the Kappa Hi-Fi HotStart ReadyMix (KAPA Biosystems: KK2601, Cape Town, South Africa). Primers for amplification were purchased from Integrated DNA Technologies (Coralville, IA, USA). The sequences for the primers were as follows: 5′ GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C 3′ and reverse primer 5′ TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG 3′ (4 nmol Ultramer
® DNA Oligo 55 bases: Integrated DNA Technologies, Coralville, IA, USA). The extraction control described above was included in the amplification, as was an amplification control (Zymo Research: D6306, Irvine, CA, USA).
Library preparation was performed according to the standard instructions of the 16S Metagenomic Sequencing Library Preparation protocol (Illumina TM, San Diego, CA, USA). This included PCR cleanup, Nextera index PCR, and quality control in the form of validation using the 4200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). The library was quantified, normalised, pooled, and denatured. Libraries were then sequenced on the MiSeq platform using the MiSeq Reagent kit v3 (Illumina, San Diego, CA, USA) and paired-end 2 × 300 bp sequencing was performed at the Sequencing Core Facility, National Institute for Communicable Diseases. The buffer-only negative control was also amplified and sequenced.
2.2. Bioinformatics
TrimGalore (v0.6.5-1) was used to filter and pair-end trim sequences to remove the Nextera adapter sequences. To ensure clean data were used for downstream analysis, quality control was performed using MultiQC (v1.6). All the downstream analyses were performed in Rstudio (v4.2.1), including classification, abundance estimations, statistical analysis, and visualisation. The Dada2 package (v1.24.0) was used for quality inspection, filtering, trimming, dereplication, sample inference, merging paired-end reads, and removal of chimeric sequences [
44]. Amplicon sequence variants (ASVs) and the ASV abundance estimates were determined using training sequence sets based on the SILVA reference database v138;
https://zenodo.org/record/4587955#.Y9JGXnZBxPY (accessed on 13 March 2024). Dada2 outputs were then constructed into phyloseq objects using the phyloseq package (v1.40.0) [
45]. The phyloseq objects were then used for further analyses.
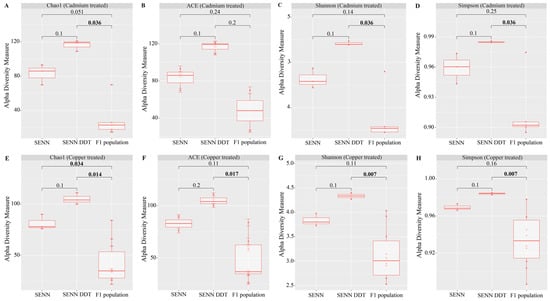
Alpha diversity is a measure of the diversity within a sample. This can be measured as the number of species present (species richness) or the abundance and distribution of species (species diversity).
For alpha diversity, species richness was determined using the Chao1 and ACE index while diversity was measured using the Shannon–Weiner and Simpson dominance index. The Wilcoxon rank-sum tests were used to compare alpha diversity between strains (or treatments).
Beta diversity is a difference in diversity between samples. For beta diversity, ordination plots were constructed using the non-metric multidimensional scaling (NMDS) method. The data clustering between the different strains for each metal treatment for the NMDS plot was statistically assessed using a PERMANOVA (permutation test with pseudo-F ratios) as implemented in the adonis function in the vegan package
https://github.com/vegandevs/vegan (accessed on 13 March 2024). To visualise overlapping microbial communities between the different families, genera and species, UpSet plots were generated using Venn Diagram (v1.7.3) and UpsetR (v1.4.0) [
46]. Differential abundance analysis between sample groups was performed using DESeq2 (v1.24.0) [
47]. Except for the UpSet plots, all plots were constructed using ggplot2 (v3.4.0) [
48].
4. Discussion
The larval environment is an important determinant of adult microbiota. However, despite increasing reports of adaptation of the An. gambiae complex adapting to breeding in polluted water, there is not a large body of data about the effect of these pollutants on the gut bacterial composition.
When comparing the overall alpha diversity of adults treated with metal as larvae, there was no significant difference from that of their untreated counterparts. However, within the metal treatments, there were small differences between the three groups. Cadmium treatment resulted in differences in species richness and diversity between the F
1 population and SENN-DDT. This was similar to the copper treatment, although the Chao 1 richness index indicated a decrease in the richness of the F
1 population compared to both strains unlike in the cadmium treatment. The lack of a marked difference in alpha diversity between treated and untreated samples is not unprecedented as it was also found in Pygmy grasshoppers that bred in mining areas [
49]. Alpha diversity in copper-treated Chironomids did not differ from their untreated counterparts at lower concentrations but decreased at higher concentrations [
50]. This suggests that the lack of differences seen in this experiment was potentially due to low-concentration exposure, as the concentrations used were at the acceptable concentration threshold for these metals [
31].
Despite the lack of variation in alpha diversity, there was a marked difference in beta diversity after treatments. As expected, the F
1 population had a greater beta diversity than that of the laboratory strains. While the metal-treated laboratory strains clustered together with their untreated counterparts, they did not overlap with the F
1 population. The F
1 population, by contrast, had a large overlap between the three groups. This suggests that while the F
1 population had a greater diversity, the laboratory strains had a more distinctive bacterial diversity after treatment compared to the F
1 population. There is not a large body of data to compare this to, but it is worth noting that copper exposure in Chironomids resulted in a similar beta diversity pattern [
50].
A key aim of this study was to determine whether larval metal treatment of laboratory strains differing in insecticide-resistant phenotype and geographical origin to an F
1 population from South Africa had similar bacterial dynamics in response to treatment. This would provide information on the utility of laboratory strains in understanding environmental effects on bacterial dynamics. This study indicated that there were large-scale differences between all three strains examined. For SENN, both metal treatments resulted in more unique genera represented (copper 15, cadmium 14, compared to 2 untreated). For SENN-DDT, the number of unique genera did not differ greatly (cadmium 10, untreated 8, copper 7). The F
1 population, however, had 43 unique genera in response to copper treatment, which was markedly more than the 16 unique genera in the untreated group and only 5 in the cadmium-treated group. Despite these differences, the F
1 population was more similar in response patterns to SENN than SENN-DDT. This suggests a role for the insecticide-resistant phenotype. The unselected SENN strain has low-level insecticide resistance and is more comparable in resistant phenotype to the F
1 population [
51] than SENN-DDT. This suggests that the selection for insecticide resistance may alter the dynamics of the bacterial response to polluted larval environments.
When comparing the genera between the three groups in response to treatment, the F
1 population consistently had the greatest number of unique genera. By contrast, SENN always had the least number of unique genera. SENN and SENN-DDT always had more shared genera than either of the strains with the F
1 population. There were very few genera shared between the three groups (3 in the untreated, 5 in the cadmium treatment and 12 in the copper treatment). This suggests a poorly conserved core microbiome shared between the three populations. This is consistent with previous studies that indicated clustering of mosquito microbiota by sampling location [
52]. This is related to habitat-specific microbiota with geography driving biodiversity differences [
53]. As such, the current study provides more information about patterns of responses rather than details about specific changes in the symbiont composition.
An examination of the differentially abundant genera shows a marked difference in response to metal treatment. In the SENN and SENN-DDT strains, the shared genera made up the greatest proportion of genera. This is reflected in the differential abundance, where the significantly abundant genera in the metal-treated individuals were similar in number to that of the untreated group. Furthermore, there were many shared genera differentially abundant in both treated and untreated groups. By contrast, there were many more differentially abundant genera in untreated F
1 adults compared to the metal-treated groups. This is noteworthy due to the high numbers of unique genera in the F
1 population, particularly in the copper-treated group. Yet, very few were differentially abundant. Copper is a bacterial micronutrient [
54] and essential for insect physiology [
55], and this could explain why copper treatment frequently resulted in the greatest number of unique genera and had fewer negative effects on the mosquito. This was true for SENN and the F
1 population. For SENN-DDT, the number of unique genera did not differ greatly in any of the treatments. This again reinforces a link between the bacterial response and the insecticide-resistant phenotype. Crucially, the metal treatment, particularly in the F
1 population, therefore increased the number of genera present, but this change in abundance was not significant. The effect of metal exposure on gut microbial composition has not been well examined in insects. Exposure to environmental metal decreases the diversity of gut microbiota vertebrates such as tree sparrows [
56]. The diversity of gut microbiota was decreased in the Pygmy grasshopper
Eucriotettix oculatus while increasing pathogenic bacteria [
49]. Exposure to high metal concentrations reduced the abundance of dominant bacterial families in the black soldier fly
Hermetia illucens. However, certain families were increased. These included
Brucellaceae,
Enterobacteriaceae,
Alcaligenaceae,
Campylobacteraceae, and
Enterococcaceae [
57]. Cadmium exposure decreased probiotic bacteria in the gut of the gypsy moth
Lymantria dispar. This included
Weissella,
Aeromonas, and
Serratia. Pathogenic bacteria, however, increased in abundance (
Stenotrophomonas,
Gardnerella,
Cutibacterium, Pluralibacter, and
Tsukamurella) [
58]. None of the examples, however, are aquatic, like mosquito larvae. Chironomids are aquatic insects with a notable tolerance to pollutants. In the case of metal exposure here, specific genera were increased in response to specific metals.
Yersinia, Dysgonomonas, Delftia, and
Enterococcus were increased in response to chromium treatment.
Yersinia and
Acinetobacter were increased in response to copper treatment [
50].
The most obvious suggestion is that the change in bacterial abundance was related to tolerating the metal pollutant. This was substantiated in the laboratory strains. The differentially abundant strains unique to the metal treatment tended to be more likely to be associated with metal tolerance. These included
Novosphingobium [
59],
Azospirillum [
60],
Blastocatella [
61],
Phenylobacterium [
62], and
Sphingobacterium [
63] in cadmium-treated SENN and
Pelomonas [
64],
Chryseobacterium [
65],
Ancylobacter [
66],
Spirosoma [
67],
Azospirilium [
68],
Rhodopseudomonas [
69],
Phenylobacterium [
62], and
Shinella [
70] in copper-treated SENN. In SENN-DDT cadmium-treated adults, the metal-tolerant genera
Shinella [
71],
Methylophilus [
72],
Paracoccus [
73], and
Caulobacter [
74] were uniquely relatively abundant. Similarly,
Paracoccus,
Brevundimonas [
75],
Ochrobactrum [
76],
Acidovorax [
77],
Stenotrophomonas [
78],
Mycobacterium [
79], and
Yersinia [
80] were uniquely relatively abundant in copper-treated adults. Although genera associated with tolerance to metal were relatively abundant in both treated and untreated individuals, there were more metal-tolerant genera unique to the metal-treated individuals. This would suggest a role for these bacteria in surviving metal treatment. This, however, was not as clearly demonstrated in the F
1 population where very few genera were uniquely relatively abundant. What was notable, however, was that despite the reduced number of relatively abundant genera, every single uniquely abundant genus in the metal treatment was associated with metal tolerance. For cadmium treatment, this was the genera
Lactococcus [
81],
Rahnella [
82], and
Morganella. This is similar in copper-treated F
1, where of seven uniquely relatively abundant genera in the metal-treated adults, four (
Kluyvera [
83],
Acetobacter [
84],
Rahnella [
82], and
Gluconobacter [
85]) were associated with metal tolerance.
What this suggests is that gut bacteria may be associated with the capacity to survive larval exposure to metal. This is more marked in the laboratory strains, but there is evidence that this happens in the F
1 population as well. This is not unprecedented. There are variations in life history traits in laboratory strains and wild mosquitoes. This includes blood-feeding duration, oviposition behaviour, mating success, and swarming behaviour [
86]. This is quite notable in the immune response. Insecticide-resistant
Culex pipiens differed in phenoloxidase activity compared to susceptible counterparts. However, this was not present in wild populations [
87]. Another study noted a decrease in immune function in insecticide-resistant
Cx. pipiens, but again, this was not observed in their wild counterparts [
88]. The association between larval exposure to metals and the relative abundance of metal-tolerant genera unique to metal treatment occurs in both laboratory strains and F
1 populations. Even though the effect is more marked in the laboratory strains, it is present in the F
1 adults as well. As such, it is likely that the effect occurs in the wild as well, although it is exaggerated in laboratory strains.
There is therefore a precedent for larval gut microbiota to protect from metal toxicants. However, the key question in this study relates to larval exposure to metals affecting mining for paratransgenesis candidates. In SENN, only one
Plasmodium protective genus (
Enterobacter [
89]) was uniquely differentially abundant in the cadmium-treated individuals. Two were uniquely differentially abundant in untreated individuals compared to their cadmium-treated individuals (
Bradyrhizobium [
90],
Aeromonas [
91]), while the remaining protective genera were abundant in both untreated and cadmium-treated individuals (
Acinetobacter [
91],
Bosea [
90]). In copper-treated individuals, the
Plasmodium protective genera
Sphingobacterium [
92] and
Elizabethkingia [
93] were more abundant than in their untreated counterparts. In the untreated group,
Bradyrhizobium was the only protective genus uniquely differentially abundant compared to its copper-treated counterparts.
Aeromonas and
Bosea were abundant in both copper-treated and untreated SENN. This was in contrast to SENN-DDT where four genera were more abundant in untreated individuals compared to the cadmium-treated individuals (
Delftia [
94],
Pseudomonas [
95],
Comamonas [
96], and
Elizabethkingia). There were no
Plasmodium protective genera uniquely relatively abundant in the cadmium treatment. This was similar to the response after copper treatment where
Pseudomonas,
Elizabethkingia,
Comamonas, and
Aeromonas were uniquely relatively abundant in the untreated group and there were none in the copper-treated group. The response in the F
1 group was similar to that of SENN-DDT. There were more
Plasmodium protective genera in the untreated group uniquely relatively abundant compared to the cadmium-treated group (
Delftia,
Pseudomonas,
Elizabethkingia,
Bacillus [
97]), while there were none uniquely relatively abundant in the cadmium-treated group. In the copper treatment,
Asaia [
98] was the only
Plasmodium protective genus uniquely relatively abundant in the treated group. By contrast,
Listeria [
99],
Delftia,
Pseudomonas,
Elizabethkingia, and
Bacillus were protective genera uniquely relatively abundant in the untreated group. It is also worth noting, however, that
Gluconobacter, a genus uniquely relatively abundant in both copper- and cadmium-treated F
1 adults, is associated with increased activity of the
imd immune pathway in
Drosophila [
100]. This immunological pathway is critical for protection against
Plasmodium [
101]. It is therefore worth noting that in the wild population, metal treatment reduced the amount of
Plasmodium protective genera that were relatively abundant. This is something that needs to be considered when trying to mine native protective symbionts.
It has been observed that adapting to breeding in polluted environments has acted as a selection pressure for insecticide resistance [
25,
27]. This has also been demonstrated in SENN and SENN-DDT, where copper and cadmium exposure increased deltamethrin tolerance, even in the offspring of these adults that bred in clean water [
34,
35]. It would therefore be interesting to consider whether potentially pesticide-degrading bacteria play a role in this selection. Genera that have previously been associated with pesticide-degrading bacteria were found relatively abundant in both treated and untreated SENN for both metals. Only
Sphingobacterium [
102] was uniquely relatively abundant in cadmium-treated SENN. In the copper-treated SENN,
Elizabethkingia [
103],
Chryseobacterium [
104,
105],
Rhodospseudomonas [
106],
Pelomonas [
107],
Phenylobacterium [
108], and
Ancylobacter [
109] were uniquely relatively abundant compared to only
Bradyrhizobium [
110] in the untreated group. The pesticide-degrading genera
Microbacterium [
111],
Variovorax [
112,
113],
Acidovorax [
114], and
Aeromonas [
115] were found relatively abundant in both untreated and copper-treated samples. This was less marked in SENN-DDT, where only
Camelimonas [
116],
Paracoccus [
117],
Methylophilus [
111], and
Lacunisphaera [
118] were uniquely relatively abundant in the copper-treated individuals, while eight pesticide-degrading genera were unique to their untreated counterparts [
104].
Sphingobacterium and
Microbacterium were pesticide-degrading genera that were relatively abundant in copper-treated and untreated SENN-DDT. For copper-treated SENN-DDT, the degrading genera
Paracoccus [
117],
Brevundimonas [
119],
Ochrobactrum [
59],
Acidovorax [
120],
Mycobacterium [
121],
Stenotrophomonas [
122], and
Yersinia [
123] were uniquely relatively abundant in the treated group. Only two pesticide-degrading genera were uniquely relatively abundant in the untreated group compared to the copper-treated group, and five genera associated with resistance were abundant in both groups (
Sphingobacterium [
102],
Microbacterium,
Bradyrhizobium, and
Delftia [
124]). This suggests a pesticide-degrading bacteria may potentially be enriched by larval metal treatment, and this was particularly marked for copper treatment. This was not reflected in the F
1 population, where although pesticide-degrading genera were present, they were often found in the untreated group or in both groups. For the cadmium-treated F
1, only
Morganella [
125] and
Lactococcus [
126] were uniquely relatively abundant in the treated group. This was similar in the copper-treated group where only
Asaia [
127] and
Kluyvera [
120] were uniquely differentially abundant in the treated group. This suggests that in the wild population, metal treatment is not as strong a selection pressure for pesticide-degrading bacteria. This is another example of an exaggerated response in laboratory strains that are not replicated in the wild population. The role of metal selection on the abundance of pesticide-degrading bacteria in the F1 population would need to be confirmed by examining the change in abundance of subsequent generations.
This study does have a range of limitations. It could have been improved by comparing the F1 population to laboratory strains of closer geographical locations. Furthermore, to fully characterise the role of gut bacteria in the wild population, this would need to be followed by a few generations of breeding in polluted water. Secondly, to fully confirm the protective role of bacteria against larval metal exposure, bacteria in larvae need to be analysed. What has been observed in this study are the changes that occur in adults after larval exposure. It must also be noted that associations between the genera are the responses based on the literature. Therefore, although the relevant genera may previously have been associated with metal tolerance, insecticide resistance, and protection against Plasmodium, it is not guaranteed that this will be a direct translation. Crucially, it would be important to identify the relevant genera to species and strains. This would be essential for the establishment of paratransgenesis as an intervention strategy.